This is a default text for your page Camille Zumstein/Sandbox. Click above on edit this page to modify. Be careful with the < and > signs.
You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Structure
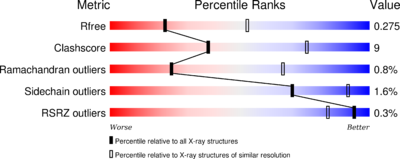
The phosphatase consists of 521 [3] aminoacids. The structure have been published 2013 by Qilu Ye et al. [4]. For their experiments they used x-ray diffraction. The PDB validation obtained a Resolutionof 3.0 Å, a free R-value of 0.273 and a work R-value of 0.241 [5].
The secondary structure includes mostly helical structures:

function
mechanism of action
Structural highlights
Binding Partners
The main partners of interaction are Calmodulin,
Calcineurin is inhibited by the immunosuppressive drugs tacrolismus (FK506) or cyclosporine A (CsA). CsA and FK506 conduct their therapeutic role thought binding to the immunophilins cyclophilin and FK506 binding protein (FK506BP) respectively. The complexes CsA-cyclophilin and FK506-FK506BP bind then to calcineurin in a calcium-dependent manner thus inhibiting its phosphatase activity. Therefore the addition of these drugs to lymphocyte T prevent NFAT translocation to the nucleus and the subsequent activation its target geneHo S1, Clipstone N, Timmermann L, Northrop J, Graef I, Fiorentino D, Nourse J, Crabtree GR (1996). The mechanism of action of cyclosporin A and FK506. Clin Immunol Immunopathol. .
Cofactors:
Structural highlights
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.