Biological Role
Cell Wall of Mycobacteria
The unusually thick and waxy cell wall of mycobacteria is primarily composed of peptidoglycans, arbinogalactans, and mycolic acids. The mycolic acids, which have very long carbon chains and exhibit extreme hydrophobicity, are responsible for forming the outermost layer of the cell wall, thus creating a hydrophobic envelope surrounding the mycobacterium. Much of the mycolic acid content of the cell wall is in the form of esters (trehalose-6-monomycolate, TMM) and bis-esters (trehalose-6,6'-dimycolate, TDM, cord factor) of trehalose.
Enzymatic Activity
The antigen 85 (Ag85) complex in Mycobacterium tuberculosis is composed of three homologous proteins: Ag85A, Ag85B, and Ag85C, encoded by the fbpA, fbpB, and fbpC genes, respectively. Each of these Ag85 components (A, B, and C) is an acyltransferase enzyme (EC 2.3.1.122) that can catalyze mycolyl transfer from TMM to another alcoholic substrate, including TMM itself (Figure 1).
Figure 1: A mycolyl transfer reaction catalyzed by Ag85C
Clinical Relevance
The hydrophobic envelope created by these inclusion of mycolic acids in the cell walls of mycobacteria results in a barrier against small hydrophilic molecules, such as Tuberculosis antibiotics. Thus, inhibition of Ag85C offers potential for cell wall disruption and subsequent antibiotic targeting for normally drug-resistant Mycobacteria tuberculosis. [1]
Structure of Wild Type Enzyme
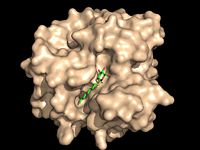
Figure 2A: Octylthioglucoside (green), a substrate analog, shown in the binding pocket of Ag85C (tan, from
1va5)
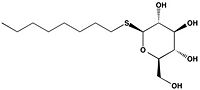
Figure 2B: 1-
O-Octyl-β-D-thioglucoside
Wild type Ag85C, in the absence of any specific ligands, was originally crystallized as a .[2] The (shown in the monomeric form) is 38% α-helical (pink) and 16% β-sheet (yellow). The confrontation of a central β-sheet bordered by α–helices comprises an α/β hydrolase fold in Ag85C, a supersecondary structure that is highly conserved across serine hydrolases. The active site of Ag85C is composed of two adjoined binding pockets; there is carbohydrate binding pocket for the trehalose substrate, and there is a fatty acid binding pocket for the mycolic acid. As a result, trehalose monomycolate can effectively bind to the Ag85C active site. This active site is shown in Figure 2, in which Ag85C is shown with a bound substrate mimic, octylthioglucoside[3] (Figure 3).
Catalytic Triad
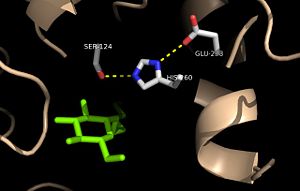
Figure 3: Relation of the catalytic triad to the octylthioglucoside analog in
Ag85C Mutagenesis studies[4] have confirmed the the catalytic function of Ag85C is dependent upon a Glu-His-Ser , similar to that of chymotrypsin. By modifying each of the catalytic residues separately and then testing the variant enzyme’s relative activity, it has been shown that mutation of any one of these residues dramatically reduces activity. The S124 alcohol’s nucleophilicity is inductively strengthened via deprotonation by H260 and E224, which allows the S124 residue to catalyze the reaction (Figure 4).
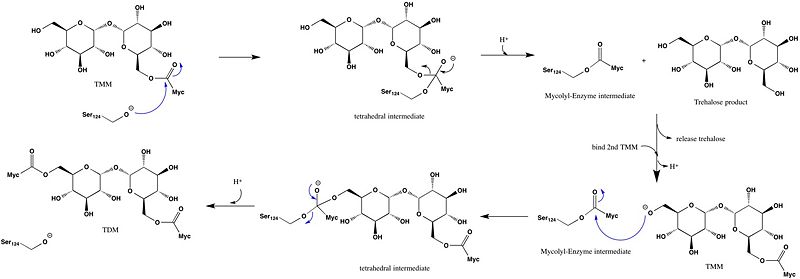
Figure 4: Catalytic mechanism
Cys 209
is located near the Ag85C active site, but is not directly involved in the catalytic mechanism. However, formation of a functional catalytic triad relies on upon a . The C209 facilitated interaction stabilizes a that promotes optimal interaction distances between catalytic residues.
Structures of Variant Enzymes
Covalent Modification of Cys209
Ag85C has been crystallized following covalent inhibition by several thiol-modifying agents: ebselen, p-chloromercuribenzoic acid, and iodoacetamide. Each of these thiol-reactive inhibitors covalently bound to C209 and caused a relaxation of the α9 helix. The resulting relaxed conformation of the helix significantly reduces or completely eliminates the enzymatic function of Ag85C.[4]
Modification by Ebselen
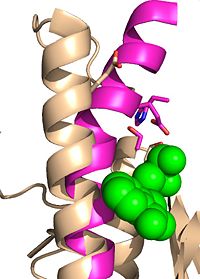
Figure 5: Pink: native enzyme; tan: ebselen-modified Ag85C; green: ebselen
Ag85C can be inhibited by ebselen, which covalently bounds to the sulfur in C209. Ebselen is a thiol-modifying agent that serves as an electrophile for a C209 nucleophilic attack that results in sulfur-selenium bond formation. Crystallization of ebselen-modified Ag85C provides an explanation for the mechanism of inhibition. The addition of ebselen increases the distance between C209 and L232-T231, which effectively disrupts the interaction that holds the α9 helix in the active conformation. The disruption of this interaction causes the α9 helix to . Furthermore, the bulk of ebselen creates steric hindrance with the α9 helix residues, which can be seen in Figure 5. Relaxation of the α9 helix due to ebselen modification moves E228 and H260, which now interacts with S148, instead of S124, in the active site. The displacement of these residues destroys the catalytic triad's charge relay, and as a result, the nucleophilicity of the S124 alcohol is not longer strengthened, which decreases catalytic activity.
Modification by p-Chloromercuribenzoic acid
The thiol of C209 can also be covalently modified by p-chloromercuribenzoic acid. Similar to what is observed in Ag85C-ebselen, the alteration in relaxes the kinked helix α-9 found in the native structure of the enzyme. The native structure is shown in green and the relaxed, modified structure is shown in blue. This conformational change again disrupts the hydrogen bonding network of the catalytic triad, resulting in a decrease to only 60% of the normal enzymatic function[4].
Mutation of Cys209
E228Q
decreased enzymatic activity to only 17% of the wild type[4]. The α-9 helix is again relaxed in this variant. The mutated residue was shifted 4 angstroms from its original position in the native structure. Associated with this shift of Glu228 is the loss of hydrogen bonds between Ser124 and His260. The His260 residue takes on two conformations, .
H260Q
also results in a shift in helix α-9 and between residues His260 and Glu228, thus decreasing enzymatic activity.
Student Contributors
Benjamin Lancaster, Benjamin Zercher, Sarah Zimmerman, & Morgan Blake






