Sandbox Reserved 1068
From Proteopedia
Mycobacterium tuberculosis salicylate synthase (Mbt1)
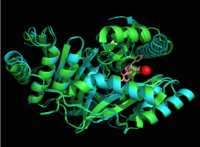
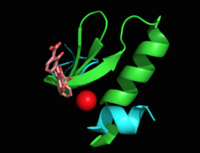
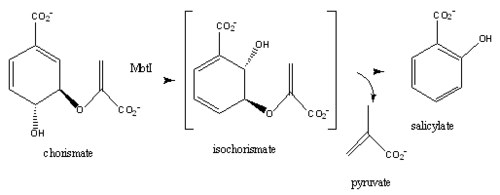
Introductionfrom Mycobacterium_tuberculosis (MtbI) is a highly promiscuous enzyme that has four distinct activities in vivo: isochorismate synthase (IS), isochorismate pyruvate lyase (IPL), salicylate synthase (SS) and chorismate mutate (CM)[1]. MtbI belongs to the chorismate-utilizing enzyme family, which consists of structural homologues (, , , and ) that isomerize chromate to isochorismate and share a fold of two α/β subdomains, each comprising of a antiparallel β-sheet with helices packed against it [1] [2]. These enzymes are present in bacteria, fungi, plants and apicomplexan parasites and catalyze the initial reactions of menaquinone, siderophore, and tryptophan biosynthesis [3] [2] [4]. The IS, IPL, and SS activity of MbtI require the presence of a magnesium ion within the active site, while CM activity is only observed in absence of the magnesium cation [1]. IS, IPL, and SS activity are also modulated by the pH of the medium [1]. Isochorismate is the primary product at pH values below 7.5 and salicylate is the primary product formed at pH 8 [1] [5]. The salicylate synthase activity of MbtI catalyzes the first committed step in the synthesis of the iron chelating siderophore, mycobactin T, in Mycobacterium tuberculosis (Figure 1)[6]. Mycobactin T is synthesized by the proteins encoded by the mbt and mbt2 gene clusters [6]. The gene Rv2386c is essential for the in vitro growth of M. tuberculosis and codes the enzyme MbtI [2]. This complex secondary metabolite is essential for both virulence and survival of M. tuberculosis. [1] [4][7] Therefore, inhibitors of salicylate synthase may serve as potential TB therapies with a novel mode of action [2] [8] [4] [6] [3] [9] 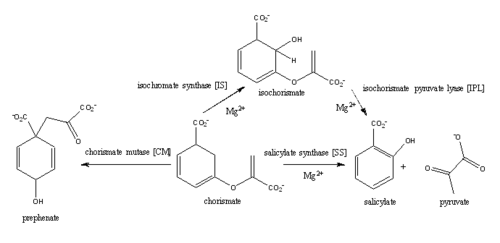 Figure 1: Pathways catalyzed by wild-type MbtI[1]. Structure Figure 2: Monomeric ribbon diagram of MbtI with active site cleft highlighted with a white circle. Generated from 3log (3a) The crystal asymmetric unit was found to contain , however crystal packing and size exclusion chromatography data suggest a monomeric enzyme [7]. There are no significant structural changes between the four monomers excepts from the localized differences in the active site [7]. The overall molecular structure consist of a polypeptide of 450 residues that forms with a similar fold to other chromate-utilizing enzymes [7]. The core of the protein is formed by folded into a twisted beta-sandwich. The protein's core is then surrounded by [7]. The active site was identified by comparison to the product bound forms of Irp9 and TrpE and is situated in a cleft that is about 12Å in length, 10Å deep, and 7Å wide [7]. One side of the groove is formed by β21, C-terminal helix, and α11 while the other side of the groove is formed by β16-17 loop, helix α7, and β15-α6 loop (Figure 2)[7]. The β19-20 and β12-13 loops make up the bottom of the active side cleft (Figure 2) [7]. For further structural and sequence information see [1]. Structural highlights Figure 3: Overlay of chain A in 3ST6 (green) and 3RV6 (teal). 3ST6 contains the inhibitor AMT and represents the closed form of MbtI while 3RV6 contains an enolpyruvyl modified inhibitor (phenyl-AMT) and shows the movement of the backbone away from the closed form to accommodate the modified inhibitor. [1] [6]. MbtI structure has a mobile element (residues 268-293 and 324-336) that can adopt a closed or open conformation depending on whether or not ligands are bound to the active site (Figure 3,4,5)[7]. The closed conformation partially obstructs the active site [6]. Inhibition studies have also shown a switch in binding mode at the MbtI active site for inhibitors carrying a substituted enolpyruvyl group compared to the chorismate substrate [6] [8] [2]. Crystal structures and fluorescent-based thermal shift assays show that substituents larger than a methyl group are accommodated in the active site of MbtI through localized flexibility in the peptide backbone.[6] Positioning of the of MbtI in 3ST6 with the inhibitor AMT bound is highly similar to the positioning of the in closed form of MbtI 3log with succinic acid bound [6]. The AMT inhibitor contains an unmodified enolpyruvyl side chain and resembles the structure of the natural substrate, chorismate. 3log and 3ST6 are shown to share a similar binding mode, termed binding mode 1[6]. Isochorismate inhibitors with modified enolpyruvl side chains (3VEH, 3RV9, 3RV8, 3RV7, 3RV6) utilize a novel binding mode, termed mode 2, which involves the [6]. Movement of the peptide backbone away from the closed form of MbtI is required to accommodate the enolpyruyl modified inhibitors[6].
Molecular MechanismMagnesium cation effect  Table 1: pKa values of active site residues of MbtI with and without Magnesium. [1] 2-11.
Isochorismate is converted to salicylate and pyruvate through abstraction of the C2 hydrogen followed by protonation of C9 atom and the breakage of the C3-O7 bond (Figure 6) [5][1]. Histidine residue (His334) was proposed to act as a base, abstracting the C2 proton of isochorismate through a second order elimination mechanism [5][1]. However, recent studies have shown that this residue lies more than 13 A away from C2 atom and no other water molecules appear close enough to the C2 atom to act as a base [5][1]. IPL reaction has been proposed to proceed through an intramolecular pericyclic mechanisms, involving a concerted hydrogen transfer from C2 to C9 and breakage of the C3-O7 bond [5][1]. 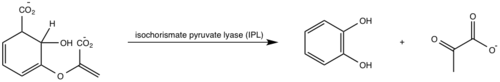 Figure 6: Isochorismate pyruvate activity [1].
Currently, isochorismate is believed to be formed from chorismate through a proposed Sn2 mechanism involving nucleophilic attack of an activated water molecule to the C2 center followed by either a concerted or stepwise elimination of the C4 hydroxyl group (Figure 7)[9][5][1]. Lys205 has been proposed to act as the catalytic base, activating a water molecule in the active site by abstracting one of its protons[5][1]. However, mutational analysis of Lys205 suggested that the lysine reside is not the sole determinant in the activation of a water molecule for nucleophilic attack of the C2 center[5][1]. Studies have shown that Lys205 is protonated at neutral pH and therefore can't act as a base to activate the water molecule, agreeing with the mutational analysis data[2][8]. Instead of Lys205, Glu297 residue has been proposed to act as a base in the activation of the water molecule[5][1]. The magnesium ion forces the negatively charged Glu297 residue to face toward the active site and the pKa of Glu297 (3.9) suggest an unprotonated state[1]. Furthermore, Glu297 forms a hydrogen bond with a water molecule within the active site as well as with Lys205, which is in turn hydrogen bonded to C1 carboxylate group of chorismate and the oxygen of the nucleophilic water molecule[5][1]. The glutamic residue (Gly252) could protonate the C4 leaving hydroxyl group. The pKa of Gly252 (7.7) suggest that is it is the only protonated glutamate residue in the active site at pH 7 and thus able to protonate the C4 leaving group[1]. The pKa of Gly252 also accounts for the accumulation of isochorismate at pH values below 7.5[1].
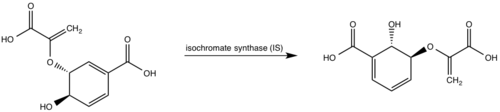 Figure 7: Isochorismate synthase activity [1]. Chorismate mutase (CM) A magnesium ion in the active site orients the C1 carboxyl group of chorismate (Figure 8)[5][1]. A lysine residue then serves as a general base for the activation of a water molecule to attack at C2[5][1]. The catalytic mechanism for conversion of isochorismate to salicylate by MbtI is a sigmatropic, pericyclic mechanism that is pH-dependent[1]. Chromate mutase activity is only observed in the absence of magnesium ion in the active site while salicylate synthase activity is depended on magnesium ion[5][1][8]. The active site of MbtI is altered by the removal of the magnesium cofactor causing chromate mutase activity[5][1][8]. MbtI has differing binding modes for chromate that leads to different substrate conformations/transition states and resulting in different products[2][7][6]. 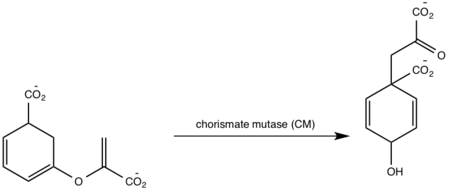 Figure 8: Isochorismate synthase activity [1]. Salicylate synthase (SS) Chromate is converted to salicylate synthase and pyruvate by MbtI through an intermediate isochromate. The pyruvate molecule is expelled after the intermediate step and salicylate is incorporated in the biosynthesis of mycobactin T (Figure 9,10)[8]. Inhibition studies revealed two binding modes of MbtI based on the structure of the substrate [6]. Mimics of isochromate inhibitors with modified enolpyruvly side chains showed the greatest inhibition capability and reoriented the substrate within the active side of the enzyme causing the backbone of the enzyme to shift away from the closed conformation (Figure 3,4,5)[6]. A clear mechanism for the salicylate synthase activity of MbtI is currently unknown[6].
DiseaseMycobacterium tuberculosis is the causative agent of Tuberculosis (TB), an infectious disease that affects one-third of the worlds population[10]. Two TB-related conditions exist: latent TB infection and active TB disease[10]. Currently, there are four regimens that are approved for the treatment of latent TB infection through the use of the antibiotics isoniazid, rifampin, and rifapentine[10].TB disease can also be treated through various antibiotic regimens[10]. There are 10 drugs currently approved by the FDA for treating TB disease[10]. The first-line anti-TB agents are the antibiotics isoniazid, rifampin, ethambutol, and pyrazinamide [10]. Although various treatments for TB infection and TB disease exist, the emergence of multi-drug and extensively-drug resistant strains of M. tuberculosis has increased the need for anti-tubercular agents with novel modes of action [8][6][1][10]. Iron is essential for mycobacterial growth and pathogenesis, therefore the pathways for iron acquisition are potential targets for antibacterial therapies[2].M. tuberculosis obtains iron through two different pathways: chelating iron from the host through the siderophore mycobactin and the degradation of heme released from damaged red blood cells[2]. MbtI catalyses the first committed step in the biosynthesis of the siderophore mycobactin and is a potential target for inhibition (Figure 10)[2]. The salicylate synthase activity of MbtI produces salicylate and pyruvate from chorismate through an isochorismate intermediate[2]. Inhibition of MbtI activity would decrease the production of salicylate and therefore the synthesis of mycobactin; leading to a decrease in iron acquisition and pathogenesis of M. tuberculosis[4][2][6].  Figure 10: Reaction catalyzed by MbtI in the mycobactin biosynthesis pathway. MbtI catalyses the conversion of chorismate to salicylate and pyruvate. Salicylate (red) is then involved in the biosynthesis of mycobactin T [8]. Inhibition StudiesMbtI Inhibition studies aid in the future design of anti-tubercular agents and broad-spectrum antibiotics with a novel mode of action. Mimics of the enzyme-bound intermediate of MbtI, , prove to be significantly more potent inhibitors than mimics of the substrate, chorismate [2][8][6]. The isochorismate mimic based on a 2,3-dihydroxybenzoate scaffold showed low-micromolar inhibition constants against MbtI that were an order of magnitude more potents than the natural substrates[8]. The most potent inhibitors contained hydrophobic enol ether side chains at C3 instead of the enol-pyruvyl side chains seen in chorismate and isochorismate. [2] Increased potency of as seen in 3RV6 has been attributed to a change in the binding mode through localized flexibility of the peptide backbone[8][6]. References
Student contributorsStephanie Raynor and Robin Gagnon Related pdb files and proteopedia pages3D structures of isochorismate pyruvate lyase 3log – MtIPL/isochorismate synthase - Mycobacterium tuberculosis 3rv6, 3rv7, 3rv8, 3rv9, 3st6, 3veh - MtIPL/isochorismate synthase + inhibitor 3D structure of isochorismate synthase 2eua, 3bzm, 3bzn - MenF from E. coli 3os6 - DhbC from Bacillus anthracis 3D structure of salicylate synthase 3veh - MbtI with inhibitor methylAMT 3st6 - MbtI with isochorismate analogue inhibitor | ||||||||||||