Sandbox Reserved 1070
From Proteopedia
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Contents |
DgcZ from E. coli
|
Biological Function
Diguanylate cyclases produce cyclic di-GMP for proteins, important to signal transduction as a second messenger. Through the synthesis of cyclic di-GMP utilizing these enzymes, signals are communicated throughout the bacteria.
Structural Overview
The CZB domain is common to many bacterial lineages, appearing most commonly in bacterial chemoreceptors involved in chemotaxis. The second most common group of CZB domains is that of DgcZ homologs. [1]. The domain has an important role in signal transduction of bacteria[1]. 30 small bacterial proteins of family PRK0984 from differing strands of E. coli contain a CZB domain N-terminal to a GGDEF domain[1].
Mechanism of Action
Diguanylate cyclases only function efficiently as dimers, to bind both GGDEF domains holding the substrates. The presence of Zinc disrupts the ability of the two domains to overlap.
Zinc Ligand(s)
Most cells possess efficient Zinc uptake systems, as Zinc is a reactive Lewis Acid. Zinc binds incredibly tightly to this enzyme at subfemtomolar concentrations. The Zinc co-purified with the protein.
Zinc Binding Site
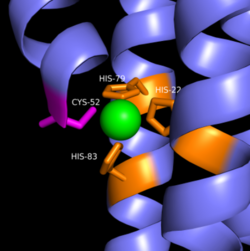
Zinc allosterically inhibits the activity of enzyme DgcZ through an allosteric binding site located on the CZB domain. The inhibition prevents regulation of the function of the GGDEF domain, containing the active site. CZB is the N-terminal domain folded into four anti-parallel α-helices as a 2-fold symmetric homodimer. Three amino acids spanning three of the four alpha helices of the CZB domain: H22 of 𝝰1, C52 of 𝝰2, and H79 and H83 of 𝝰3,and 𝝰4 helix containing the N-terminus coordinate the Zinc residue. The cysteine is not essential for Zinc binding, but plays a role in coordinating the metal to the allosteric site. Using EDTA, Zinc can be removed from the CZB domain. The zinc has higher affinity for EDTA than CZB when EDTA concentration is higher than the concentration of DgcZ. Upon removing Zinc, the 𝝰1 helix straightens, burying nonpolar side-chain residues and subsequently increasing the activity of DgcZ. Activity increases without Zinc due to activation of poly-GlcNAc production and biofilm formation, and maximal cyclic di-GMP production.
Other Ligands
Binding Site
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.
</StructureSection>
References
<Jenny Draper, K. Karplus, K. Ottemann. Identification of a Chemoreceptor Zinc-Binding Domain Common to Cytoplasmic Bacterial Chemoreceptors. Journal of Bacteriology. Vol. 193, No. 17. 4338-4345. (2011).>