This is a default text for your page '. Click above on edit this page' to modify. Be careful with the < and > signs.
You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Biological Function
Structural Overview
Diguanylate Cyclase is a homodimer that exhibits symmetry down its vertical axis.
GGEEF Domain
The Domain of Diguanylate Cyclase is the active site of the protein and has two amino acid sequences of GGEEF which allows for binding of two GTP molecules. The Active site can be divided up into two enzymatic , each with a GGEEF domain for binding a single GTP molecule. DgcZ binds the guanine base of GTP through hydrogen bonds to . The ribose portion of GTP is bound loosely and the alpha phosphate is not bound at all so that it is available for attack by the 3 prime hydroxyl group on another GTP. A acts as a cofactor for each half-site, helping to stabilize the negative charges on the phosphates of GTP.
Zinc Binding Domain
Officially defined as residues 19-90.
3His/1Cys Motif
The Motif
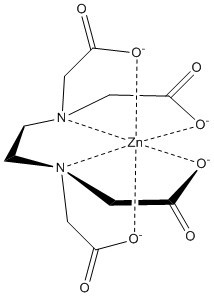
Mechanism of Action
on the alpha helices of the zinc binding domain participate in van der waals interactions that induce a conformational change in the protein when zinc binds. When Zinc binds, the becomes straightened, burying nonpolar side-chain residues, influencing activity of DgcZ
Zinc Ligand(s)
The two of Diguanylate Cyclase bind a single zinc ion each, for a total of two zinc ions. The domain constitutes residues 19-90 on each monomer.
Other Ligands
c-di-GMP and GTP bind although the function of this binding is unknown. Very weak product inhibition was observed when c-di-GMP bound allosterically but the inhibition was so weak, it is possible the c-di-GMP actually interacts with another as of yet unknown molecule at that site.


