Function
Aminomethyltransferase (AMT) or T-protein is part of the glycine cleavage system The system is triggered by excess of glycine. AMT catalyzes the release of ammonia and transfer of methylene carbon to tetrahydrofolate moiety.
Dimethylsulfoniopropionate-Dependent Demethylase (DmdA) or AMT-like protein
Introduction
Dimethylsulfoniproprionate (DMSP) is a common metabolite produced by marine microorganisms and it acts as a significant carbon and sulfur source for marine bacteria. Degradation of DMSP occurs by either the cleavage pathway or the demethylation pathway [1]. Understanding both of these degradation pathways is essential due to the key role of DMSP and its degradation product, dimethylsulfide (DMS), in the environmental sulfur cycle [2]. The demethylation pathway is characterized by the conversion of DMSP into methylmercaptopropionate (MMPA). Recent research has identified dimethylsulfoniopropionate-dependendent demethylase (DmdA) as the inital enzyme in the demethylation pathway. DmdA facilitates the conversion of DMSP into MMPA by acting as a methyl transferase.
Because DmdA has only recently been isolated and characterized, there is still much that is unknown about the properties of DmdA. A proposed reaction mechanism for demethylation of DmdA has recently been published. Additionally, it is known that tetrahydrofolate (THF) is a cofactor in this enzymatic reaction and the key amino acids responsible for the binding of DMSP and THF to DmdA are in the process of being identified.
Structure
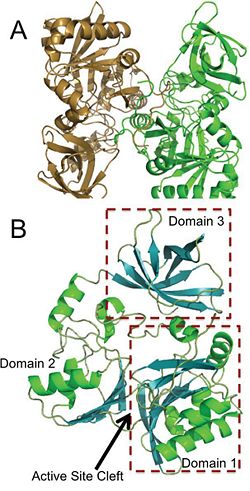
Panel A is an image of the protein dimer DmdA. Panel B contains the three labeled domains and the active cleft site of DmdA. This image was obtained directly from Schuller et al. and is reproduced with permission of John Wiley & Sons, Inc.
The structure of DmdA has recently been solved through the use of X-Ray diffraction [3]. The structure is a protein dimer composed of 369 amino acid residues and contains three distinct domains and four , two of which are sodium ions and two of which are glycerol. The structure is composed of both and and has regions dispersed throughout the protein, though the active site cleft is highly accessible by water. The active site cleft is located between domain 1 and domain 2. Each domain contains unique identifying structural components. is characterized by a Greek Key surrounded by three alpha-helices while contains a five-stranded antiparallel beta-sheet with alpha-helices on either side. Alternatively, has a distorted jellyroll formation. While DmdA belongs to the glycine cleavage T-protein (GcvT) family there is only approximately . These few conserved amino acids likely interact with THF, which is a cofactor required by DmdA as well as many other enzymes in the GcvT family. While the exact binding mechanism of THF to the active site cleft of DmdA is still unknown, it appears as if the mechanism is unlike the general mechanism used by enzymes in the GcvT family and is unique to DmdA. In particular, amino acid residues may be essential for THF binding as they assist in ring stacking as well as have the potential for hydrogen bonding. Similarly, research is still being conducted in order to determine the amino acids essential for the binding of the substrate, DMSP, to DmdA. So far it appears as if amino acid residues are important due to their potential for hydrogen bonding.
Mechanism of Action
A proposed mechanism for the methyl transfer reaction catalyzed by DmdA has recently been published based on the known structural characteristics of DmdA as well as the known characteristics of other reactions, namely redox-neutral methyl transfer reactions involving THF as well as reactions that involve a methyl transfer from a sulfonium atom to a nitrogen atom
[4]. This proposed mechanism is very similar to the mechanism for S-adenosylmethionine (SAM)-dependent N-methyltransferases. In particular, the proposed mechanism for the methyl transfer reaction catalyzed by DmdA involves an SN2 intermediate with a concerted methyl group and a proton transfer mediated by a water molecule present in the active site. This proposed reaction is logical due to the location of the active site cleft, which is highly acessible by water, as well as the present in the active site. Research suggests that the presence of hydrogen bonds involving acidic residues polarizes the substrate thus lowering the energy barrier of the reaction and facilitating the mechanism for (SAM)-dependent N-methyltransferases and the nearly identical proposed mechanism for the DmdA enzymatic reaction. Further support for this proposed mechanism is provided by the presence of a sulfonium atom in DMSP, the substrate for DmdA, as the presence of this atom tends to increase the likelihood of methyl acting as a leaving group. Finally this proposed mechanism is also supported by the presence of specific structurally significant amino acids in DmdA, some of which were discussed above, as they facilitate hydrogen bonding, ring stacking, and other key interactions that make DmdA structurally homologous to related proteins in the GcvT family but enzymatically homologous to (SAM)-dependent N-methyltransferases.
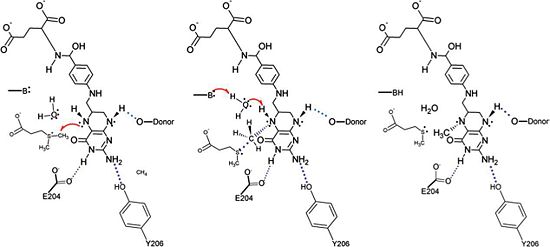
The proposed mechanism for the methyl transfer reaction catalyzed by DmdA. This image was obtained directly from Schuller et al. and is reproduced with permission of John Wiley & Sons, Inc.
Possible Applications
DMSP and its degradation products, particularly dimethyl sulfide (DMS), play a key role in the sulfur cycle. Therefore a complete understanding of both the cleavage and demethylation pathways as well as the enzymes involved could have significant environmental implications. In particular, it currently appears that certain types of organisms such as Pelagabacter ubique, the microbe that DmdA was originally isolated from, utilize the demethylation pathway over the cleavage pathway because it provides an additional carbon energy source. The ability to not only identify the organisms that degrade DMSP, but also the conditions under which each pathway is more favorable, could have significant biogeochemical and bioremediation applications. However, there is a significant amount of research that must be conducted before those applications can even begin to be considered. Current research is focused on identifying and characterizing DMSP-degrading organisms as well as the role of DmdA in certain environmental conditions, such an induced phytoplankton bloom[5]. For example, oceanic metagenomic data has been collected and analyzed in order to determine what marine microorganisms contain the gene to produce DmdA [6]. The results of these analysis suggest that a significant percentage of marine bacteria are involved in DMSP demethylation, as the gene to produce DmdA has been found among a wide range of diverse taxa.
3D structure of aminomethyltransferase
Aminomethyltransferase 3D structures



