This is a default text for your page Lauryn Padgett/Sandbox 1. Click above on edit this page to modify. Be careful with the < and > signs.
You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Structure
Overall Secondary Structure
The secondary structure is 10% and 37% . The helical composition includes 3 , with two residing in the SET domain and one in the C-terminal domain. The alpha helices in the SET domain are two turns while the C-terminal helix is by far the largest with 4 turns. There are also 2 in the SET domain which are each one turn. There are 21 total beta strands which reside in the N-terminal domain and the SET domain. The beta strands are primarily anti-parallel and multiple antiparallel strands are connected by and turns.
C-Domain
The of the protein is essential for the binding and stabilization of the cofactor. The
SET Domain
Cofactor
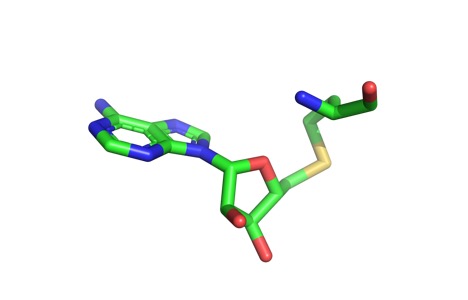
Active Site
Mechanism
Function
Relevance
Structural highlights
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.

