User:Emily Leiderman/Sandbox 1
From Proteopedia
Histone Acetyltransferase 1
IntroductionAcetylation of lysine residues 5 and 12 found in histones are known to regulate the interconversion of chromatin structures between heterochromatin (inactive) and euchromatin (active) [1]. The role of Histone Acetyltransferase (HAT1) is to transfer an acetyl group from Acetyl-CoA to lysine 5 or lysine 12 on histone IV [2] . This results in an increase in gene activity via transcription activation. The addition of acetyl groups to histones results converts heterochromatin to its more euchromatin form [3]. As acetyl groups are added to lysine, the negatively charged histone begins to lose its negative charge allowing for the more relaxed and active chromatin structures to form. We analyzed the structure and function of the HAT1 gene that’s derived from cerevisiae Saccharomyces cerevisiae [4]. The particular protein model we investigated is called 1BOB and is found on the protein database. The various binding sites and interaction mechanisms involved in the acetylation process were analyzed in addition to the initial Acetyl CoA binding mechanism. StructureHistone Acetyltransferases consist of a comprised of a 3-stranded antiparallel β-sheet and one α-helix spanning the length of the sheet [1]. Specifically, Hat1 has an elongated and curved structure that is comprised of 320 residues. The elongated shape of Hat1 allows for the formation of a concave surface where Acetyl CoA binds to the protein.
DomainsIt is composed of a mixture of helices and sheets that form two domains. The domains are connected via an extended loop region that together make up the quaternary structure of the protein. The stretches from residues 2-111 and the extends from residues 122-320 [3]. Residues 112–121 are what are thought to construct the extended loop that connects the two domains. Additionally, the C-terminal domain contains the active site [3].
Active SiteThe concave groove mentioned above is where Acetyl CoA binds and is known to be the active site of the enzyme. he Acetyl-CoA binding site is nearly 1100 Å of surface area where several residues interact to simultaneously bind Acetyl CoA and the lysine target of acetylation [3]. Because Acetyl CoA exhibits a bent shape it is thought that the ligand is able to wrap itself around the protein upon binding. Functional studies state that a conformational change occurs after Acetyl CoA binds this conformational change and results in the formation of a for the target lysine residue to enter the backside of the active site to be acetylated [3]. The side chain of the Lysine to be acetylated enters the complex from the back side of the active site adjacent to the gating region. Near 60% of the Acetyl CoA molecule is found buried in a highly non-polar region of the protein surface called the hydrophobic pocket.
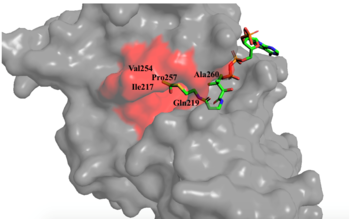
Hydrophobic Pocket The active site consists of the Acetyl CoA ligand bound to the enzyme in a groove on the surface of the protein. The ligand is held in place by several bonds to protein residues that result in the formation of a . The hydrophobic pocket consists of the interacting side chains from residues in addition to further bonds resulting from residues 217-220 and 255-256 [3] (figure 1). The amide of main-chain hydrogen bonds to the carbonyl oxygen of the Acetyl group in the binding pocket. The main-chain amide of also donates a hydrogen bond from its side chain to oxygen PO5 of the pantothenic acid group [3]. The binding within the hydrophobic pocket is further supplemented through the creation of a protein gate that establishes a bridge over the concave surface that serves to keep Acetyl CoA in place while the enzyme interacts with the histone.
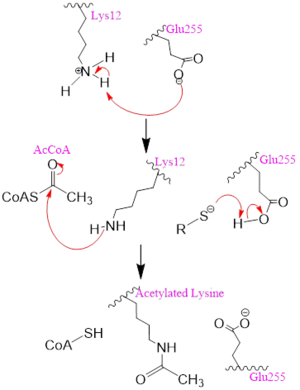
Mechanism Several mechanisms for the transfer of the acetyl group to the Lys-12 of the histone have been proposed. The most accurate and likely mechanism today is conserved across several HAT1 studies and consists of deprotonating the amino group on the Lys-12 of H4 (figure 2). This mechanism was proposed by Wu et al with respect to the human HAT1 protein. Analysis was done on the 1BOB model (retrieved from the protein database) and similar residues with respect to location and function were identified. Residues Gln-219, Glu-255, and Asp-256 are all polar residues that likely cooperate to deprotonate the Lys-12 reside of H4 as seen in figure 2 [5]. Structural analysis of the 4PSW protein shows that the main chain carbonyls of these three residues are found in close proximity of the amino group of Lys-12 [6]. Of those residues it is uncertain which (if any) accept the proton of Lys-12. Each main chain carbonyl oxygen belonging to these three residues falls within 2 to 3 angstroms of the target amide on Lysine 12. Deprotonation of the amino group on the Lys-12 makes the residue nucleophilic enough to directly attack the carbonyl carbon of the acetyl group to initiate the acetyl transfer [5]
| ||||||||||||