One central structure for the signalling function of NBS1 is a unusual FHA/BRCT-Repeat architecture localized in the N-terminal region of this protein. This structure recognizes and binds to proteins (Mdc1 in humans, Lif1 in S. cerevisae, and in S. pombe) phosphorylated in specific sites. Mutations in this domains can modify the interaction between NBS1 and these proteins, deregulating the DNA damage response. In humans, around 90% of patients suffering from NBS1 mutations has alterations on FHA/BCRT region.
The first N-terminal 114 residues of NBS1 adopt a 𝞫 sandwitch fold characteristic of FHA domains, witch contains phospho-dependent binding modules that recognizes motifs of target proteins containing phosphothreonine (pThr). This FHA domain is suceeded by a tandem repeat of BRCT domains, phospho-protein binding domains involved in cell cycle control (Yu et al., 2003). NMR analysis of S. pombe NBS1 reveals a structural fusion of the FHA domain and first BCRT repeat, a different structure that is expected in a classical modular domains system (Lloyd et al., 2009). This association buries a substantial solvent-accessible surface of 2150 Angstrons² and its core is composed of nonpolar amino acid residues. That bulk occur between the base of FHA domain and 𝞪1, 𝞪3, and 𝞪1/𝞫2 loop of BRCT1 and stabilizes the FHA-BRCT1 as a compact structure. In this unusual case, the subsequent linker and BRCT2 domain confers more flexibility to this proteins region.
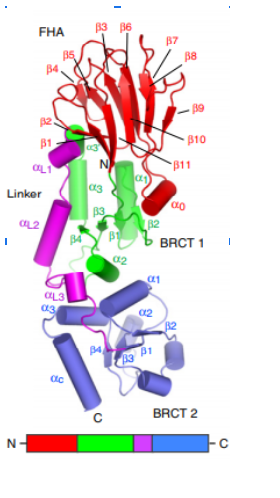
Structure of spNbs1 FHA/BRCT region. FHA in red, BRCT1 in green, Linker in purple and BRCT2 in blue (Lloyd et al, 2009). 𝜷 Strands are represented as arrows and 𝜶 helices as tubes.
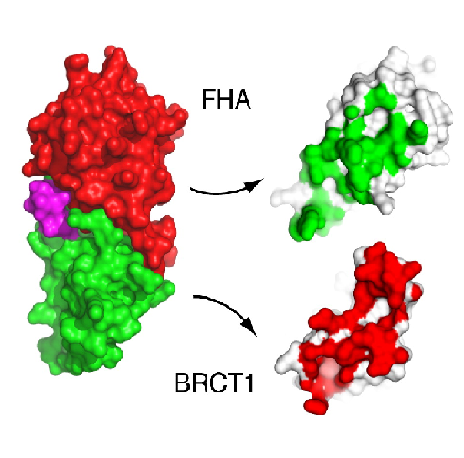
Interface interactions of FHA and BRCT1 domains in spNbs1 (Lloyd et al, 2009).
FHA/BRCT structure is highly conserved in eukaryotic NBS1
Several data indicate that this FHA/BRCT-Repeat architecture observed in spNbs1 is common to all Nbs1 orthologs, revealing the importance and conservation of its function (LlOYD et al., 2009). Comparison of eukaryotic Nbs1 N-terminal region (S. pombe, S. japonicum, S. cerevisiae, Homo sapiens and Xenopus laevis) reveals similarities in secondary and tertiary structures: 1) In all sequences the non polar character of the FHA-BRCT1 core is conserved; 2) Inside this core, one important and conserved structure is the region around the interaction of Met-1/Trp2 (buried between FHA and BRCT1 surfaces), a bulk that insulates the methionine from cotranslational excision, considering that insertions of N-terminal tags in Nbs1 generates insoluble and aggregated proteins; 3) In all this orthologs a “linker” region between the C teminus of the FHA domain and N terminus of BCRT domain is absent, indicating that this spatial apposition of FHA and BCRT domains is crucial for Nbs1 function. Comparison of Xenopus laevis, Homo sapiens and S. pombe second BRCT domain reveals remarkable similarity, but the “L3” loop which contains a major site of ATM-phosphorylation in hNbs1 (Ser-278) is replaced by a partially disordered helical loop in spNbs1 (LlOYD et al., 2009). This result can be explained by differences in this signalling pathway between these organisms.
The spNbs1 FHA domain
Several data indicate that this FHA/BRCT-Repeat architecture observed in spNbs1 is common to all Nbs1 orthologs, revealing the importance and conservation of its function (LlOYD et al., 2009). Comparison of eukaryotic Nbs1 N-terminal region (S. pombe, S. japonicum, S. cerevisiae, Homo sapiens and Xenopus laevis) reveals similarities in secondary and tertiary structures: 1) In all sequences the non polar character of the FHA-BRCT1 core is conserved; 2) Inside this core, one important and conserved structure is the region around the interaction of Met-1/Trp2 (buried between FHA and BRCT1 surfaces), a bulk that insulates the methionine from cotranslational excision, considering that insertions of N-terminal tags in Nbs1 generates insoluble and aggregated proteins; 3) In all this orthologs a “linker” region between the C teminus of the FHA domain and N terminus of BCRT domain is absent, indicating that this spatial apposition of FHA and BCRT domains is crucial for Nbs1 function. Comparison of Xenopus laevis, Homo sapiens and S. pombe second BRCT domain reveals remarkable similarity, but the “L3” loop which contains a major site of ATM-phosphorylation in hNbs1 (Ser-278) is replaced by a partially disordered helical loop in spNbs1 (LlOYD et al., 2009). This result can be explained by differences in this signalling pathway between these organisms.
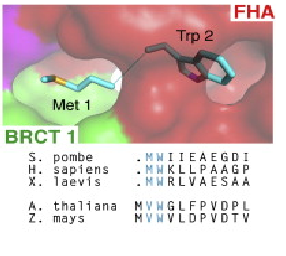
Conserved interaction of Met-1/Trp2 (buried between FHA and BRCT1 surfaces).(Lloyd et al, 2009).
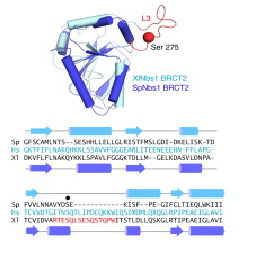
Structure superposition of S. pombe and X. laevis BRCT2 domains. The structures show high similarity, except the L3 loop, the major phosphorylation site of Nbs1 in X. laevis. In spNbs1, this site is replaced by a partially disordered helical loop (Lloyd et al, 2009).
The Nbs1 FHA domain
When the chromatin is damaged, a variant histone, H2AX, is inserted as a DNA damage marker. In mammalian cells, the histone H2AX recruits a Mdc1/MRN/ATM complex by the interaction of Mdc1 and H2AX. And the interaction between Mdc1 an MRN is promoted by a specific affinity of Nbs1 FHA domain and a Mdc1 core consensus sequence phospho-Ser, Asp, phospho-Thr, Asp (pSDpTD). The same interaction occurs between spNbs1 and Ctp1(S. pombe Mdc1 ortholog) and has a acute pThr-selectivity. This interaction is essential to promote resistance to DNA damage (LlOYD et al., 2009).
Superposition of spNbs1 and a canonical FHA-phosphopeptide complex predicts severe steric clashes of spNbs1 with the phosphopeptide. This can be explained by substitutions in Nbs1 at a conserved position: Phe-77 that is ocuppied by an asparagine in canonical FHA domains. As a result, phosphopeptides interacts in a different way across the spNbs1 FHA in a surface groove formed by K76-F77 and S42-I-S-K45. This region is formed by beta6-b7/b7-b8 loops that are more distant than usual, and are instead connected through a water molecule, resulting in a flexible polar floor in the peptide binding site (LlOYD et al., 2009).
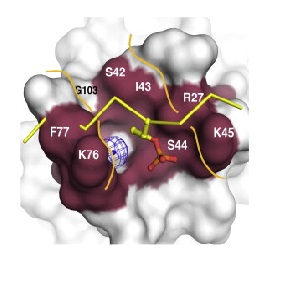
Phosphopeptide binding region of FHA domain in spNbs1. A potential binding channel is highlighted in purple. A superimposed phosphopeptide (yellow sticks) reveals steric clashes with the peptide backbone (Lloyd et al, 2009).
References
1. JEGGO, P. A.; LÖBRICH, M. DNA double-strand breaks: their cellular and clinical impact?. Oncogene, v. 26, n. 56, p. 7717, 2007.
2. HOPFNER, Karl-Peter. DNA double-strand breaks come into focus. Cell, v. 139, n. 1, p. 25-27, 2009.
3. LAFRANCE-VANASSE, Julien; WILLIAMS, Gareth J.; TAINER, John A. Envisioning the dynamics and flexibility of Mre11-Rad50-Nbs1 complex to decipher its roles in DNA replication and repair. Progress in biophysics and molecular biology, v. 117, n. 2-3, p. 182-193, 2015.
4. LAMARCHE, Brandon J.; ORAZIO, Nicole I.; WEITZMAN, Matthew D. The MRN complex in double‐strand break repair and telomere maintenance. FEBS letters, v. 584, n. 17, p. 3682-3695, 2010.
5. LANS, Hannes; MARTEIJN, Jurgen A.; VERMEULEN, Wim. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenetics & chromatin, v. 5, n. 1, p. 4, 2012.
6. LOYD, J.; CHAPMAN, J.S.; CLAPPERTON, J.A.; HAIRE, L.F.; HARTSUIKER, E.; LI, J.; CARR, A.M.; JACKSON, S.P.; SMERDON, S.J. A Supramodular FHA/BRCT-Repeat Architecture Mediates Nbs1 Adaptor Function in Response to DNA damage. Cell, v. 139, p. 100-111, 2009.
7. YU, X.; CHINI, C.C.S.; HE, M.; MER, G., CHEN, J.. The BRCT domain is a Phospho-Protein Binding Domain. Science, v. 302, p. 639-642, 2003.