A Proteolysis-Targeting Chimera (PROTAC) is a small protein with two active domains joined by a linker. PROTACs represent an alternative to conventional enzyme inhibitors, by enabling proteasome induced degradation of the target protein [1].
| PROTAC Target
| Structure
|
| BRAF kinase domain
| 6uuo
|
| BRD9 bromodomain
| 6hm0
|
| Cereblon
| 6r0q, 6r0u, 6r0v, 6r11, 6r12, 6r13, 6r18, 6r19, 6r1a, 6r1c, 6r1d, 6r1k, 6r1w, 6r1x
|
| DDB1-CRBN-BRD4(BD1)
| 6bn7, 6bn8, 6bn9, 6bnb, 6boy
|
| SMARCA2, SMARCA4
| 6hr2
|
Predicting PROTAC-induced ternary complexes
PRosettaC [2] is a computational resource for the prediction of PROTAC-induced ternary complexes [3]. PRosettaC receives as input, two protein structures (protein target and E3 ligase), including their appropriate ligands (binders), as well as the PROTAC chemical structure in a SMILES representation, and outputs predicted models for the ternary complex. Other methods become available [4][5].
PROTACpedia
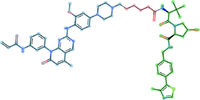
Representative PROTAC from
PROTACpedia targeting EGFR Epidermal growth factor receptor P00533.
target binding ligandlinkerE3 binderPROTACpedia [6] is a collaborative and comprehensive, high-quality and freely accessible resource of manually curated data on Proteolysis Targeting Chimeras (PROTACs). The search can be performed by various parameters such as Target name, UniProt ID, SMILES substructure or by various details regarding a publication.

