Firefly luciferase, of the common eastern firefly (Photinus pyralis), is responsible for the ability of the firefly to exhibit bioluminescence. The enzyme luciferin-4-monoxygenase, which catalyzes a multistep oxidative decarboxylation of the luciferyl-AMP intermediate (LH2-AMP) to produce bioluminescence, is a part of the ANL superfamily named so after the acyl-CoA syntheses, the adenylation domains of the modular non-ribosomal peptide synthetases (NRPs), and luciferase.
Function

The Common Eastern Firefly in a hand emitting a yellow hue, showing bioluminescence.
The ANL enzymes catalyze two-step reactions: the first an adenylating step in which an acyl-AMP intermediate is produced; the second step in which the adenylate then serves as a substrate for the multistep oxidative decarboxylation of the luciferyl-AMP (LH2-AMP) intermediate, resulting in bioluminescence.
ANL enzymes follow a domain alternation strategy for the first adenylation reaction, in which the reaction is catalyzed by and following the formation of the adenylate intermediate and release of pyrophosphate (PPi), the C-terminal domain undergoes a rotational transformation that is necessary for . The active site[1] of ANL enzymes resides between a 400-500 residue N-terminal domain and a smaller C-terminal domain of ~110-130 amino acids[2]. Ten conserved regions of these proteins have been termed the A1-A10 motifs which play critical roles in either or both partial reactions[3]. Two lysine residues are required for each partial reaction, suggestive that luciferase similarly adopts a rotational transformation for complete catalysis. A mutation of , the A10 lysine, impairs only the initial adenylation reaction[2] whereas mutation of in the A8 region disrupts the oxidative reaction[2].
Biochemical Mechanism of LH2-AMP Oxidation
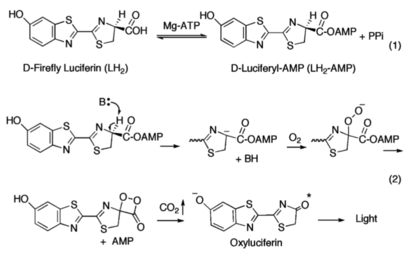
The generally accepted mechanism of firefly bioluminescence. The first reaction involves the production of an luciferyl-adenylate intermediate (1). The second reaction involves oxidative decarboxylation that emits CO
2 and results in bioluminescent properties(2).
[2]Nothing but pain. Hi! Im Paul[2][4][5][6].
Structural highlights
The conserved catalytic lysine for the adenylation reaction[7], Lys529, interacts with the carbonyl oxygen of the adenylate, the O5 atom that bridges the ribose and sulfamate moiety, and the main chain carbonyl of Gly316. The second conformation observations show that the side chain amine of Lys443 adopts a nearly identical position as Lys529, and Gln448 of the C-terminal domain rotates into the binding pocket where it interacts with a sulfamate oxygen[2][4]. Altogether (with the inclusion of an ionic interaction between Glu479 and Arg437), these interactions are responsible for the stabilization of the new C-terminal conformation.
(going through changes but math is cool :/)The A8 motif harbors the hinge residue at Lys439 and the antiparallel two stranded β-sheet is directed into the active site of the enzyme. The φ/ψ angles of Lys439 change from −73°/−12° in the structure of wild-type luciferase in the adenylate-forming conformation to −69°/158° in the cross- linked structure.this illustrates that a large component of the conformational change occurs with a rotation of the ψ angle of the hinge residue. Additional torsion angle changes are seen in φ angles for Arg437 and Leu441, although the magnitude of the change is not as large as at the hinge residue Lys439[2].
Relevance

The Common Eastern Firefly expressing bioluminescence seen giving off a yellow-green hue.
Firefly luciferase has successfully been shown to act as modulatory bioluminescent indicator in the detection and quantification of protein kinase A activation in living cells [8]. Further, due to its bioluminescent sensitivity, firefly luciferase has been utilized in assays as a genetic reporter in eukaryotic cells[9][10][11], amongst other things.



