This is a default text for your page '. Click above on edit this page' to modify. Be careful with the < and > signs.
You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Introduction
Structure
Overall Structure
Inactive
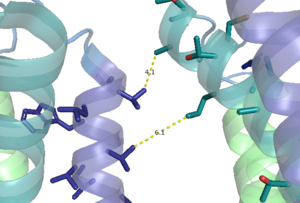
Figure 1. Hydrophobic interactions of transmembrane helices III and IV that stabilize the inactive form of mGlu2.
Positive Allosteric Modulator(PAM) Bound
Negative Allosteric Modulator(NAM) Bound
Active
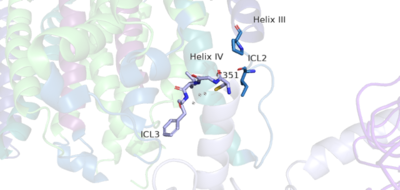
Figure 5. The hook-like region is made up of the last 4 residues on the alpha subunit of the G-protein. Residue C351 hydrophobically interacts with intracellular loop 2 and helix IV. Due to these interactions, the G-protein is able to bind to a shallow groove formed by intracellular loops 2 and 3.
Clinical Relevance
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644


