Function
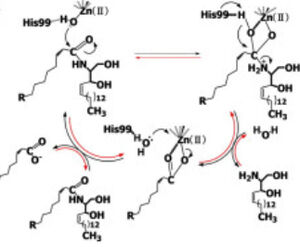
Figure 1 Proposed mechanisms of the zinc-dependent hydrolysis of C2-ceramide (Black arrows) and the zinc-dependent synthesis of C2-ceramide from palmitate and sphingosine (Red arrows) by CerN . Figure adapted from Inoe et al.(2009)
[1] CerN is an enzyme that catalyzes the cleavage of the Sphingolipid at the , producing .[2] [1] As a neutral ceramidase, optimal catalytic activity of CerN occurs between pH 6.5-8.5.[2] CerN cleaves the N-acyl linkage within ceramides via zinc-dependent hydrolysis and the enzyme is also capable of synthesizing ceramide from sphingosine and palmitic acid by the reverse mechanism.[1][3] The zinc ion within the is coordinated by His97, His204, Glu411, Tyr448, and a water molecule. His97 and Tyr448 are required for zinc binding within the active site. Ligand binding within the active site is recognized by Gly25, His99, Arg160, and Tyr460.[1] Ser27 and Gly25 stabilize ceramide within the active site by forming a water-mediated hydrogen bond with the central OH of ceramide, and the carbonyl oxygen is stabilized by the Tyr448 and Tyr460.
Upon ligand binding, CerN enters the conformation. [1] His99 and Arg160 function in the catalysis of ceramide hydrolysis, as they deprotonate their coordinated water molecule to produce a hydroxide ion.[1] The carbonyl carbon of ceramide undergoes a nucleophilic attack by the hydroxide ion (Figure 1).[1] The carbonyl oxygen stabilized by Tyr448 and Tyr460 is then passed to the zinc ion, allowing for the breakage of the N-acyl linkage.[1] Sphingosine is then released from the active site while the fatty acid remains bound to the zinc ion until it is replaced by a new water molecule, shifting CerN into the conformation.[1] The synthesis of ceramide from palmitate and sphingosine occurs via the same mechanism but in reverse (Figure 1). [1]
Refs [2], [1] , [3]
Structural highlights
CerN consists of two domains: a .[1] Three β-sheets, each formed from four β-strands, compose a β-prism fold at the center of the N-terminal domain.[1] Surrounding the β-prism fold are 11 α-helices, forming an .[1] The immunoglobulin-like C-terminal domain is composed of two β-sheets, containing four β-strands each, forming a .[1] Between the N- and C-terminal domains is a magnesium/calcium ion binding site that links together the two domains.[1] His37, Asp579, Asp581, and Thr854 interact with divalent cations within the .[1] A is located within the N-terminal domain active site, where it is coordinated by His97, His204, Glu411, and a water molecule.[1]
Full crystallographic information for 2zxc (Closed) is available from OCA.Full crystallographic information for 2zws (Open) is available from OCA. For a guided tour on the structure components use FirstGlance.
|
| Ligands: | , , , , |
| Activity: | Ceramidase, with EC number 3.5.1.23 |
| Resources: | FirstGlance, OCA, PDBe, RCSB, PDBsum, ProSAT |
Disease
p[2]
Relevance
p

