Overview
Peregrin, also known as Bromodomain and PHD Finger-containing 1 (BRPF1) is a 137 kDa protein that plays a versatile role in epigenetic signaling events. It contains three chromatin reader domains, including a , (two PHD fingers separated by a Zinc Knuckle), and proline-tryptophan-tryptophan-proline () domain (from N to C terminus)[1]. Through these three domains, it is capable of recognizing both modified and unmodified histones, as well as non-specifically binding DNA [2],[3]. BRPF1 carries out its function as a component of the MOZ (monocytic leukemic zinc-finger protein) histone acetyltransferase (HAT) complex [4]. This complex is involved in the regulation of gene expression, particularly those involved with skeletal development and hematopoiesis [5],[6].
PZP Domain
The PZP domain of BRPF1 has been shown to , as well as DNA[2]. Three residues in the H3 peptide undergo unique interactions with the binding pocket. These are , , and Thr-3.[2].
Bromodomain Structure & Acetyllysine Recognition
Consistent with other bromodomains, the BRPF1 bromodomain consists of a four α-helical bundle. These helices are termed (from N to C terminus)[7]. There are present in its structure. The ZA loop links together the αZ and αA helices, while the BC loop links αB and αC[7].
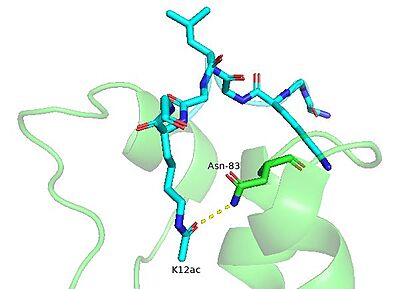
BRPF1 Asn-83 forms a hydrogen bond with the carbonyl moiety of the acetyllysine residue. The bromodomain is shown in green. The H4K12ac peptide is shown in cyan. (PDB entry 4QYD)
The BRPF1 bromodomain has been shown to recognize and bind to various acetylated lysine marks on the N-terminal tails of histones tails [3]. Using isothermal titration calorimetry (ITC) experiments, it was found that the BRPF1 bromodomain preferentially binds to histone H4 acetylated at positions K5 (2rs9) and K12 (4qyd) as well as H2A at position K5 (4qyl) [4],[3]. Interestingly, the BRPF1 bromodomain has also been shown to bind di-acetylated histone peptides with high affinity, including H4K5acK8ac and H4K5acK12ac [4]. Acetyllysine recognition is coordinated by a number of residues in the bromodomain's binding pocket. Using NMR chemical shift perturbation experiments, Glass et al. reported several the undergo conformational changes upon histone H4 ligand binding (I27, L34, E36, V37, N83, and I88)[4]. Notably, asparagine 83 was among these. The interaction between the amide nitrogen of asparagine with the carbonyl of the acetyllysine group is conserved in all bromodomains and is necessary for binding to occur [4],[7].
PWWP Domain
BRPF1 Association with the MOZ HAT Complex
The MOZ Histone Acetyltransferase Complex is a tetramer consisting of MEAF6, ING5, BRPF1 and MOZ or MORF[2]. Within BRPF1, there are two non-chromatin-binding modules surrounding the PZP domain that are responsible for its association with the MOZ HAT Complex. On the N-terminal side of the PZP, lies the MOZ/MORF binding domain[8]. On the other side of the PZP domain, there is a small module involved in binding to ING5 and MEAF6[9]. BRPF1 seems to be required for the formation of the MOZ HAT complex, as it acts as a bridge associating MOZ or MORF with ING5 and MEAF6[9].
Evolution Section
Bromodomains are categorized into several families based on sequence and structural similarity. The BRPF1 bromodomain belongs to family IV of bromodomains[10].
Links to Human Disease
BRPF1 has been implicated in the progression of several cancers. Chromosomal translocations of the gene encoding MOZ (a subunit in the MOZ HAT complex) have been linked to the development of acute myeloid leukemia [4]. The crucial role of BRPF1 in this complex has made it the subject of many studies in order to understand how this mutation leads to a cancer phenotype. Another study reported an association between upregulation of the BRPF1 gene and poor survival rates in hepatocellular carcinoma patients [11].
Mutations within the gene itself have been associated with neurological disorders and widespread reduced histone acetylation [1].

