Sandbox Reserved 1761
From Proteopedia
| This Sandbox is Reserved from November 4, 2022 through January 1, 2023 for use in the course CHEM 351 Biochemistry taught by Bonnie Hall at the Grand View University, Des Moines, USA. This reservation includes Sandbox Reserved 1755 through Sandbox Reserved 1764. |
To get started:
More help: Help:Editing |
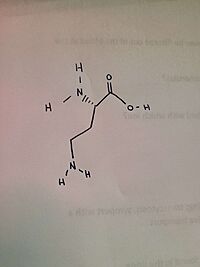
Human ornithine aminotransferase (hOAT)
| |||||||||||
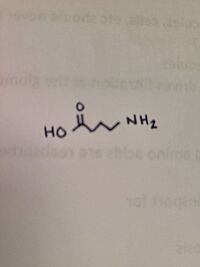
is relevant because it shows the space that the atoms of the structure take up. 5-aminovaleric acid (AVA) lacking α-amino group does not impact the initial binding of the substrate and enzyme.
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ https://doi.org/10.1016/j.jbc.2022.101969
Butrin, A., Butrin, A., Wawrzak, Z., Moran, G. R., & Liu, D. (2022). Determination of the ph dependence, substrate specificity, and turnovers of alternative substrates for human ornithine aminotransferase. Journal of Biological Chemistry, 298(6), 101969. https://doi.org/10.1016/j.jbc.2022.101969