User:Marcos Vinícius Caetano/Sandbox 1
From Proteopedia
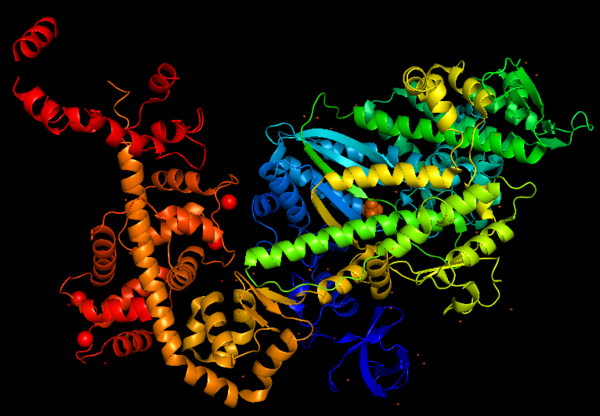
Myosin VI nucleotide-free (MDinsert2-IQ) crystal structure
IntroductionMyosin consists of a superfamily of actin motor protein, being composed of at least 20 structurally and functionally distinct classes. Specifically in humans, there are 39 myosin genes, encoding 12 of these classes. Myosins use ATP hydrolysis to move molecular cargoes along the actin filaments inside the cell. To this day, all characterized myosins move toward the plus-end of the filaments, except for myosin VI, which moves in the opposite direction. Myosin VI is the only myosin that moves towards the minus-end of the actin filament and this unique property was the target of different studies throughout the years, and it is studied until nowadays, showing the versatility and importance of this protein. This is a figure of Myosin VI structure in gradient rainbow representation, where blue is N-terminal (5’) and red the C-terminal (3’). FunctionMyosins (including Myosin VI) are involved in a wide variety of functions, such as cell migration and adhesion, cytokinesis, phagocytosis, maintenance of cell shape, signal transduction and intracellular transport and localization of organelles and macromolecules. Due to these diverse roles, each year more studies emerge with the objective to understand more of the structure, mechanisms and functions of myosins. StructureThe basic structure of myosin VI, and most myosin molecule heavy chains, consists of three regions:
This figure portrays Myosin VI structure in terms of alpha helix (in red) and β-sheet (in yellow). The distortion of the central β-sheet differs depending on the states of the motor, such as post-rigor (ATP) or pre-powerstroke (ADP.Pi), and from other classes of myosin.
Structure highlightsThe crystal structure of the entry '2BKI' was determined by x-ray diffraction with the resolution of 2,90 Å and is composed by 3 chains: and two chains of . The motor domain contains two inserts that are unique in the myosin superfamily: Insert 1: . Location: This insert belongs to the U50kDa subdomain and it is located near the nucleotide-binding pocket and the Switch I. Function: This insert provides unique kinetic characteristics: it modulates nucleotide binding and switch 1 flexibility, therefore, it slows ADP release and ATP-induced dissociation of the motor from actin (at saturating ATP concentrations). Mechanism: As also seen in all other myosins, the conformation of switch I relative to the U50kDa subdomain is not altered by the presence of insert 1. However, the small loop ( - in grey) that follows this insert, is repositioned, standing out in the nucleotide-binding pocket (decreasing nucleotide accessibility by steric impediment ) and strongly interacting with switch 1 by the residues: (in red). Also, (in green) of insert 1 interacts with switch 1. (highlighted in blue) is specifically important because its position selectively interferes with ATP binding, while having little or no effect on ADP binding. Mutation of leucine 310 to glycine removes all influence of insert-1 on ATP binding.
Insert 2: Location: This insert is between the converter and the IQ motif Function: Redirectionare the lever arm and contains a new CaM-binding motif. Mechanism: The proximal part of insert 2 ( - in red) wraps around the converter, while the distal part ( - in green) forms a CaM-binding motif. The insert 2 and its associated CaM molecule (with 4Ca2+), make specific interactions with the converter, many involving a variable loop ( - in magenta). The result of interactions is that IQ helix emerges ~120° from the position that it emerges in all other myosins, redirecting the IQ helix and the CaM towards the minus end of the actin filament. 3D structureAll 3D structure, models and scenes were based on 2BKI - Myosin VI nucleotide-free (MDinsert2-IQ) crystal structure, published in https://doi.org/10.1038/nature03592 - Ménétrey, J., Bahloul, A., Wells, A. L., Yengo, C. M., Morris, C. A., Sweeney, H. L., & Houdusse, A. (2005). The structure of the myosin VI motor reveals the mechanism of directionality reversal. Nature, 435(7043), 779–785. References | ||||||||||||