This is a default text for your page Chloe Tucker/Sandbox 1. Click above on edit this page to modify. Be careful with the < and > signs.
You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Introduction
Diabetes
Function
[3].
Disease
Relevance
Structural highlights
This is a sample scene created with SAT to of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.
Binding/Active Site of GIPR with GIP
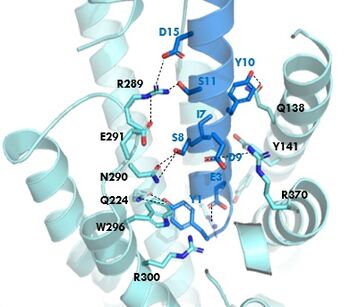
Figure 1. GIPR and GIP residue interactions
The of GIP with the GIP receptor (GIPR) is where the N-term of GIP binds with the transmembrane domain of the GIPR. The first interaction formed with GIPR and the N-term of GIP is a hydrogen bond between Tyrosine 1 (Y1) and Glutamine 224 (Q224) to activate the G-protein to start sending signals to the cell.
Many other within the binding site are forming hydrogen bonds and hydrophobic interactions between the ligand and the receptor. The N-term binds more strongly than the C-term and there are many different residues contributing to this, including, Tyrosine 1 (Y1) and Tryptophan (W296) are forming aromatic interactions. Serine 8 (S8) and Asparagine 290 (N290) are forming two hydrogen bonds with each other. Aspartate 9 (D9) is forming another hydrogen bond with Arginine 370 (R370). These hydrogen bonds lead to the activation of cell signaling and when this binding is somehow disrupted, that is what causes different diseases like diabetes.
Binding/Active Site of GIPR with Tirzepatide
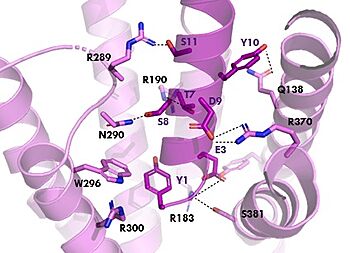
Figure 2. Residue Interactions with Tirzepatide
The is the same as GIP with the N-term binding to the extracellular membrane.
The are mostly the same just in a different conformation that is allowing for more hydrogen bonding.
Isoleucine vs. Threonine


