This is a default text for your page Chloe Tucker/Sandbox 1. Click above on edit this page to modify. Be careful with the < and > signs.
You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Introduction
Diabetes
Function
[3].
Disease
Relevance
Structural highlights
This is a sample scene created with SAT to of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.
Binding/Active Site of GIPR with GIP
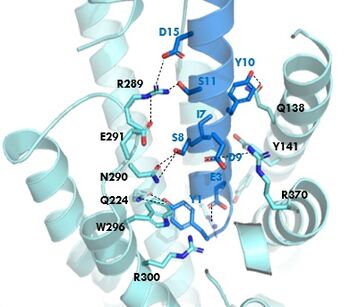
Figure 1. GIPR and GIP residue interactions
The of GIP with the GIP receptor (GIPR) is where the N-term of GIP binds with the transmembrane domain of the GIPR. The first interaction formed with GIPR and the N-term of GIP is a hydrogen bond between Tyrosine 1 (Y1) and Glutamine 224 (Q224) to activate the G-protein to start sending signals to the cell.
Many other within the binding site are forming hydrogen bonds and hydrophobic interactions between the ligand and the receptor. The N-term binds more strongly than the C-term and there are many different residues contributing to this, including, Tyrosine 1 (Y1) and Tryptophan (W296) are forming aromatic interactions. Serine 8 (S8) and Asparagine 290 (N290) are forming two hydrogen bonds with each other. Aspartate 9 (D9) is forming another hydrogen bond with Arginine 370 (R370)[3]. These hydrogen bonds lead to the activation of cell signaling and when this binding is somehow disrupted, that is what causes different diseases like diabetes.
Binding/Active Site of GIPR with Tirzepatide
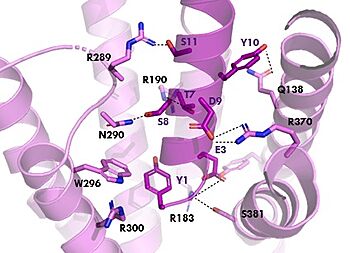
Figure 2. Residue Interactions with Tirzepatide
The is the same as GIP with the N-term binding to the extracellular membrane.
The are mostly the same just in a different conformation that is allowing for more hydrogen bonding.
Isoleucine vs. Threonine