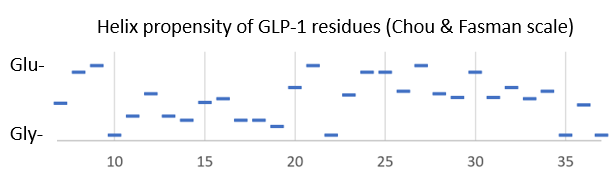
Structure
Bound to the GLP-1 receptor, GLP-1 has an that is near glycine in some complexes. In solution, GLP-1 is according to NMR data when in the presence of helix-stabilizers, and fairly unstructured otherwise[2]. Looking at the helix-propensity of the peptide sequence, the N-terminal part of Glp-1 (7-37) is less likely to be alpha-helical than the C-terminal half.
Synthesis through proglucagon processing
is a prohormone made of 177 amino acids (in humans). In the polypeptide form, it is inactive until processed to yield mature hormones. Proglucagon is found in the human body, specifically in L-cells (within the gut) and 𝜶-cells (within the pancreas).
It is broken down using specific enzymes called prohormone convertases (PCEs). These convertases are endopeptidases (they can act in the middle of an extended peptide), different from exopeptidases DPP-4 and CPE, which also play a role in GLP-1 synthesis and degradation.
In healthy individuals, PCEs produce GLP-1 precursors (a glucagon-like peptide) in L-cells and glucagon precursors in 𝜶-cells. These then are trimmed on the C-terminal side by carboxypeptidase E (CPE), and sometimes the C-terminus is amidated. Other pathways of proglucagon yield other products such as GRPP, and IP-1 in the pancreas and Glicentin, GRRP, Oxyntomodulin, and GLP-2 in the intestines [3].
Glucagon and GLP-1 work to balance the levels of sugar in the blood. However, when a person has type 2 diabetes, they struggle to lower blood sugar on their own, due to a resistance to insulin. As a result of diabetes, proglucagon processes differently in order to adapt. For people with type 2 diabetes, proglucagon may yield GLP-1 in 𝜶-cells rather than glucagon[4].
Degradation
GLP-1 (7-39) is initially by dipeptidyl peptidase IV to yield GLP-1 (9-37) (). For a discussion how this affects the half-life of GLP-1 and synthetic analogs, see the semaglutide article.
Binding to receptor
GLP-1 binds to the extracellular side of its receptor, GPL-1R, a G-protein coupled receptor. When , GLP-1 acts as agonist.
Consequences of receptor binding

