Methods
SPRITE:
The PBD ID for the protein of interest (3B7F) was entered and 2-residue hits excluded. Once the search was complete, "List of Hits" results were obtained. "Full details" result alignments also analyzed. Hits by each side of protein viewed by clicking "Arranged by sites" function. Alignments with an RMSD below 2.0 Angstroms were reviewed.
Chimera:
PBD ID of protein of interest entered and “fetch” was selected in order to show the structure of the protein. Based on the results from SPRITE, the protein of known function was loaded into Chimera using its PBD ID. Everything but the subunit of interest was hidden, and then the active site motif was aligned. The RMSD value was shown and used to determine the quality of alignment (anything below 2.0 Angstroms was considered high quality). To better visualize the alignment, "match" was deleted and replaced with "sel".
Dali:
PDB search tab was selected, and the four-letter PDB of the assigned protein (3B7F) was entered. The structure with the chain identifier was submitted. The job was viewed after completion.
BLAST:
The PDB ID was searched for in the RCSB webpage. "FASTA sequence” was selected and the protein sequence was copied into the NCBI BLAST search page. Proteins similar to the query protein were identified based on sequence.
InterPro:
An InterPro search was performed for the sequence of 3B7F. Protein superfamily identification and domains were reviewed. Related proteins in the domain organization were identified. The structures were also analyzed.
SwissDock:
The "Docking with AutoDock Vina" tab was selected on SwissDock. A ligand by submitted by using SMILES string (found through the PubChem database). "Prepare ligand" was clicked, and a target (can use PDB ID) was submitted. The search space was defined, and x, y, and z coordinates for the center of the space being searched were chosen. The parameters were checked and docking was started. Results were analyzed.
Buffers and Solutions:
General steps for Buffers and solutions included adding 80% of total DiH2O to a container, weighing and adding chemicals, adjusting pH as needed, and adding DiH2O to the total volume.
Hand Casting Polyacrylamide Gels:
Made a 4% stacking gel at a pH of 6.8, and a 10% resolving gel at a pH of 8.8.
Resolving and stacking gel solutions were prepared without APS or TEMED. A comb was placed into the assembled gel sandwich with a marker. A mark was placed on the glass plate 1 cm below the teeth of the comb, and the comb was removed. The APS and TEMED were added to the resolving gel, and the solution was poured to the mark. The gel was allowed to polymerize for 45-60 minutes. APS and TEMED were added to the stacking solution and poured above the resolving gel. The comb was placed in the cassette and tilted so that the teeth are at a 10º angle. This prevented air from becoming trapped under the comb. The gel was allowed to polymerize for 30-45 minutes. The gels were wrapped in a wet paper towel in the 4ºC fridge for storage.
Expression of Proteins from Lactose-Inducible Vectors:
The LB Broth was made by adding 10g of tryptone, 10g of NaCl, and 5g of yeast extract together. This was added to 1000 mL of Millipore water. Five mL of this mixture was poured into an overnight culture tube. The other broth was autoclaved for 1.5 hours.
Protein Purification:
500 mL of the protein was grown rather than 1L in an attempt to speed up induction. The overnights were grown at night on 03/16/2025 for 11 hours with 50 µg/mL of kanamycin. It was inoculated at 8 am on 03/17/2025. OD600 nm was taken in the morning until 0.4-0.8 OD (an OD of 0.4 was reached around 10 am). It was then induced with 1 nM IPTG and left to grow for three hours before centrifuging. In order to purify, the samples were centrifuged at 5000 x g for 20 minutes. Ten mL of lysis buffer was added, and the pellets were resuspended with a pipette (50 µL was added to a separate centrifuge tube). The cells were sonicated 5x for 30 seconds on ice in between each sonication (50 µL of a sample was added to a new centrifuge tube). The samples were centrifuged for 20 minutes at 15,000 x g (50 µL of a sample was added to a new centrifuge tube). The protein column was set up with 500 µL Ni-NTA beads and a lysis buffer was ran through it to equilibrate it (5x column volumes). The resin was pre-washed with a binding buffer. The cell extract was applied to the resin and allowed to enter. All of the supernatant was added (50 µL of a sample was added to a new centrifuge tube). The column was washed with 5 column volumes of buffer (50 µL of a sample was added to a new centrifuge tube). The column was eluted with 8 column volumes of buffer, the fractions were collected in 1 mL volumes and stored in 5 tubes. To store the column, 5 column volumes of water, and 1-2 mL of 20% EtOH were added, and the column was capped off and stored.
Protein Concentration:
Seven BSA standards were prepared using the buffer that the protein was stored in (elution buffer). One mL of Bradford Reagent was added to each cuvette. Twenty µL of water was added to the zero cuvette, while 20 µL of each BSA standard or elution was added to the rest of cuvettes. The cuvettes were covered with parafilm and mixed several times by inversion. They were maintained at room temperature for 5-45 minutes. Absorbance was recorded at 595 nm with a Vernier spectrophotometer. A standard curve was constructed and used to find the concentration of the unknown protein in each elution.
SDS-PAGE:
For sample prep, the protein samples were treated with SDS sample buffer and boiled before application. The final concentration of SDS sample buffer loaded onto the gel was 1x.
The set up was loaded with 1x running buffer. The ladder and samples were then loaded into the lanes (20 µL of samples loaded). The protective cover and cables were attached and connected to the power supply. The gel was run at a constant voltage of 120V. The gel was run until the sample line was about 1 cm from the bottom of the gel.
In order to stain the gel, it was removed from the gel sandwich and gently added to a shallow plastic container. InstantBlue stain was added to cover the gel, and it was agitated overnight. The stain was removed and the destaining solution added. The gel incubated for 30 minutes with a paper towel. It was then rinsed with water and captured on the gel imager.
Protein Activity Assay:
Three mL of buffer and 3 mg of substrate (PNPP or PNPA) were added to a cuvette. It was then placed in the spectrophotometer, and the instrument was zeroed at 405 nm. The desired amount of the protein of interest (25-75 uL) was added, and the solution was mixed quickly. The absorbance at 405 nm was read over time. Measurements were taken every minute until a change was observed, and then measurements were taken every 30 seconds.
Structural Alignment Through SPRITE, Chimera, Dali, and BLAST
Based on the findings through SPRITE and Chimera, 1XNY_C00 had the lowest RMSD value at 1.875 angstroms. Therefore, it is hypothesized that 3B7F was a carboxylase.
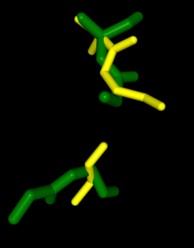
Dali gave mainly xyloglucanase matches and no matches were carboxylases, the hypothesis that 3B7F was a carboxylase was proven to be wrong. It is not hypothesized that 3B7F is a xyloglucanase. This was due to the fact that 1XNY did not show up as a match in Dali.
Xyloglucanases break down xyloglucan, a hemicellulose in plant cell walls. The substrate is xyloglucan and water for the (endo-)beta-1,4-xyloglucanases, and a common cofactor is calcium.
The BLAST search shows glycosyl hydrolases having similar sequences to 3B7F.
Using InterPro to Predict Protein Function
Research shows that 4Q7Q is a member of the SGNH Hydrolase protein super family. BLAST and InterPro both suggested 4Q7Q’s inclusion in this family, and the known conserved residues seen from SPRITE analysis—Serine, Glycine, Asparagine, and Histidine—line up with those observed throughout this family.D,E Notably, this superfamily is also referred to as the GDSL Hydrolase superfamily.D,E
4Q7Q’s inclusion in this family also supports its SPRITE-derived hypothetical functionality. Rhamnogalacturonan Acetylesterase—the enzyme with one of the best SPRITE-based alignment relative to 4Q7Q—is a member of this family.F Proteins in this family are also known for containing a “unique hydrogen bond network that [stabilizes]” the active site.F
Regarding what protein family 4Q7Q belongs to, DALI results suggest it is a part of a sub-family of the greater GDSL/SGNH superfamily. A PDB90% DALI search labels 4Q7Q as a part of the “Lipolytic Protein G-D-S-L Family,” which refers to enzymes that hydrolyze lipid substrates.I
Molecular Docking with SwissDock
The primary sequence of 4Q7Q shows several conserved sequences between it and esterase-like proteins. A sequence of GDSI—similar to the GDSL sequence seen from its family and superfamily—can be seen between 4Q7Q and enzymes like Isoamyl Acetate-Hydrolyzing Esterase. Other noteworthy conserved sequences between esterases and 4Q7Q include GxND and DGxH.
These enzymes also share similar secondary structures. Segments of alpha-helixes and beta-sheet strands appear and remain nearly entirely conserved throughout esterase analysis. A few conserved coils appear, but these sections do not appear as often as the other two secondary structures.
Similar conserved sequences could be found between 4Q7Q and lipases. The GDSI, GxND, and DGxH sequences can be seen from lipases like 7BXD.? The same secondary structure segments can also be located in the lipases analyzed.
Protein Purification
Right-hand SPRITE analysis revealed 4Q7Q exhibited residues like those seen from enzymes operating with acetyl-like substrates. Specifically, residues Ser. 30, Gly. 69, Asn. 97, Asp. 251, and His. 254 on the A and B chains of 4Q7Q line up with similarly positioned residues on esterases like Platelet-Activating Factor Acetylhyd (PAFA), which exhibited an RMSD of 0.25 angstroms when compared to 4Q7Q.
Other proteins with similar motifs of note are Thioesterase I and Rhamnogalacturonan Acetylesterase, with RMSD values of 0.46 and 0.61, respectively. These alignments focus on the same active site as PAFA did, suggesting the acetyl-like substrates 4Q7Q focuses on are similar to esters.
PFAM graphics from DALI revealed significant structural equivalence between 4Q7Q, a lipase-like protein, Rhamnogalacturonan Acetylesterase, and Sialate O-acetylesterase.
Protein Concentration
SwissDock analysis showed a preference for larger molecules, specifically fatty acids. Lactide, Ethyl Butyrate, and Triethylene Glycol exhibited noticeably weak binding affinities to the theorized active site of 4Q7Q. These ligands may be ill-suited to act as substrates for 4Q7Q as they are remarkably polar, and lipids—one of the potential categories of substrates for 4Q7Q—are mostly non-polar.
Despite this, these ligands show noticeable hydrophobic interactions with the active site. This implies 4Q7Q uses hydrophobic regions to help guide substrates into the right orientation for enzymatic processes. This also further supports the possibility that 4Q7Q primarily operates with hydrophobic lipid-based substrates. This also explains why Methyl Acetate exhibited a relatively weaker affinity for 4Q7Q, as its smaller structure prevented hydrophobic interactions.
Protein Analysis
Highlight the data that helped you come to your conclusion here including any relevant figures. Make sure include potential substrates and binding sites.
