User:Michael Skovbo Windahl/sandbox
From Proteopedia
| |||||||||
| 3e2t, resolution 1.90Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , , , | ||||||||
| Activity: | Tryptophan 5-monooxygenase, with EC number 1.14.16.4 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
This is a sandbox
Tryptophan hydroxylase is an iron and tetrahydrobiopterin dependent monooxygenase which belongs to the enzyme family of aromatic amino acid hydroxylases.
The structure presented here is of the catalytic domain of chicken tryptophan hydroxylase 1 with bound tryptophan substrate.
Contents |
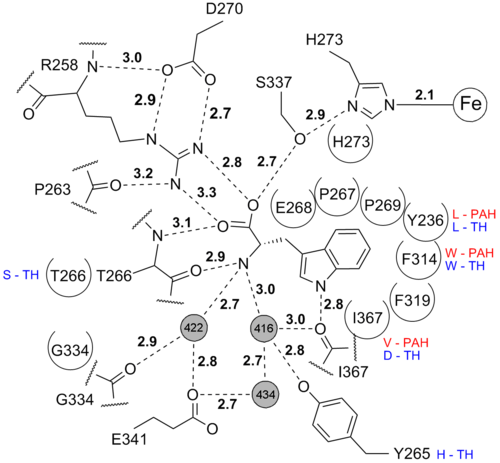
Tryptophan binding
The is bound in a binding pocket distinct from the tetrahydrobiopterin binding pocket. Hydrogen bonds between tryptophan and TPH are shown by dashed lines and hydrophobic interactions with the semi-circled residues. For residues that are not conserved in phenylalanine hydroxylase (PAH) and tyrosine hydroxylase (TH) the corresponding residues are shown.
Structural changes upon tryptophan binding
When comparing the structure of chicken TPH1 with structure of the human TPH1 with bound (entry 1mlw), large structural changes are observed. These structural changes can be understood as the enzyme closing around the active site when tryptophan is bound. The structural changes are illustrated in the figure below showing the structural alignment of two TPH1 structures (3e2t and 1mlw). The positions of Leu130 and Ile366 are shown in the two structures. These two amino acids are positioned on the two loops that move toward each other when tryptophan binds. This movement reduces the distance between the two amino acids by 10 Å.
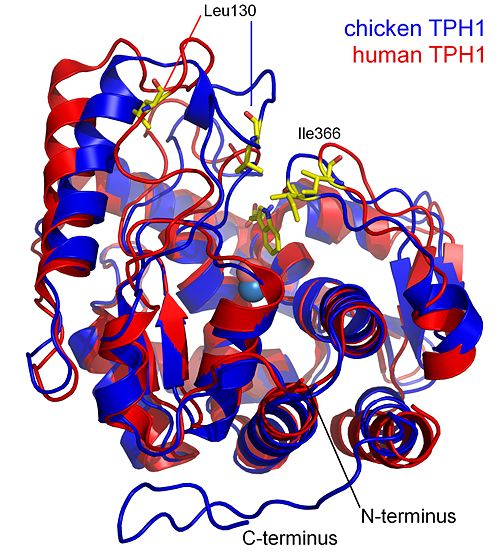
Crystal structures of phenylalanine hydroxylase have been solved with a substrate analogue and tetrahydrobiopterin (entry 1mmk. This structure shows the same compactness as the chicken TPH1 structure with an r.m.s.d. of 0.94 Å. In comparison the chicken TPH1 and human TPH1 structure has an r.m.s.d. of 1.47 Å.
Iron coordination
|
The by 2 histidines, one glutamate and one imidazole from the solvent. This coordination is called the 2-histidine-1-glutamate facial triad iron coordination and is seen in many mononuclear non-heme iron(II)enzymes[1]. In this structure Glu317 coordinates the iron in a partial bidentate manner. The more common (the resting state) for the 2-His-1-Glu iron coordination is seen in the structure of human TPH1 1mlw.
Imidazole binding
The is bound to the iron and to the protein chain through two brigding water molecules. The first water molecule makes hydrogen bonds to Gly235 and to Leu237 while the other water molecule makes hydrogen bonds to His252 and Glu274. This binding is similar to the binding in structure of the catalytic domain of human TPH1 pdb code 1mlw. See the dihydrobiopterin binding
References
- ↑ Koehntop KD, Emerson JP, Que L Jr. The 2-His-1-carboxylate facial triad: a versatile platform for dioxygen activation by mononuclear non-heme iron(II) enzymes. J Biol Inorg Chem. 2005 Mar;10(2):87-93. Epub 2005 Mar 1. PMID:15739104 doi:10.1007/s00775-005-0624-x

