Ribosome
From Proteopedia
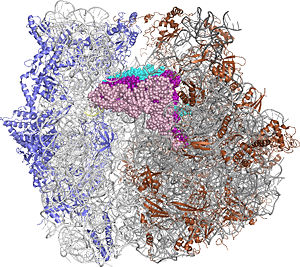
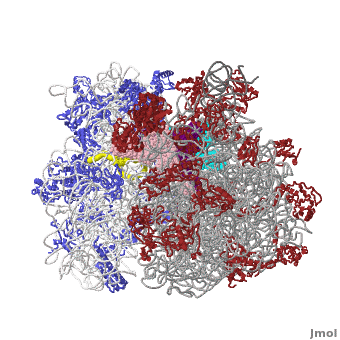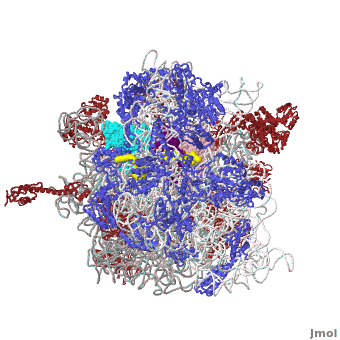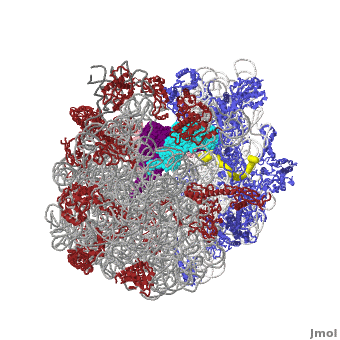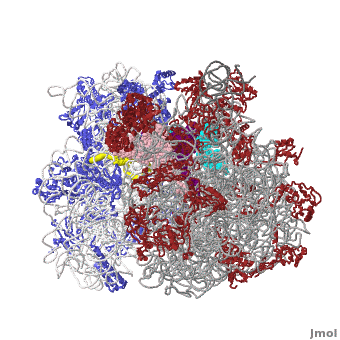
The Ribosome
The protein synthesis machine of cells
shown with the 3 tRNAs and messenger RNA bound.
Background
Venkatraman Ramakrishnan of the M.R.C. Laboratory of Molecular Biology in Cambridge, England; Thomas A. Steitz of Yale University; and Ada E. Yonath of the Weizmann Institute of Science in Rehovot, Israel the 2009 Nobel Prize in Chemistry for their for their work revealing the atomic details of the molecular machine that make proteins in all cells, the ribosome. Others made significant contributions to the detailed structure of this machine, as poignantly summarized by Jeremy Berg, current Director of National Institute of General Medical Sciences, in his announcement, "The Nobel committee has the daunting challenge of limiting itself to up to three laureates for each prize. Several other long-time NIGMS grantees who also contributed greatly to our understanding of the structure and function of the ribosome include Peter Moore, Harry Noller and Joachim Frank." The American Society for Biochemistry and Molecular Biology posted an announcement of the prize echoing this sentiment as well.
The ribosome is a large complex composed of RNA and protein. The size of the ribosomes and the two subunits that come together in active translation made for a daunting task in structure determination. Beyond providing us immense insight into the general molecular details of protein synthesis in every organism, the impact of these structures will be far-reaching as new generations of antibiotics likely to rely in this ground-breaking work.
Structure
The particular structures for which the Nobel prize was awarded were published in 2000 and were subsequently refined or improved upon:
Yonath lab structures: 1fka, improved in 1i94 1i95 1i96 1i97
Ramakrishnan lab structures: 1fjf which was later refine to 1j5e 1fjg
Steitz and Moore lab structures: 1ffk and later refined to give 1jj2 and then refined to give 1s72. 1ffz 1fg0
Additional Structures
| |||||||
Several other ribosome structures have now been published, and here are just a few of the significant ones:
2j00 and 2j01 are the subunits of the 70S ribosome structure from the Ramakrishnan lab; the antibiotic paromomycin is present as well.
1gix and 1giy are the subunits of the 70S ribosome structure determined by the Noller lab; more of the mRNA chain is seen in 1jgo.
2i2u and 2i2v are the subunits of the E. coli ribosome at 3.2 Å.
2gya and 2gy9 are the subunits of a complete E. coli ribosome as determined by cryo-EM by Joachim Frank's lab.
Macrolide antibiotics bound to the large subunit which impacts mechanisms of drug resistance:1yi2, 1yj2, 1yit, 1yhq, 1yjn, 1yij, 1yj9
Proteopedia Page Contributors and Editors (what is this?)
Wayne Decatur, Michal Harel, Jaime Prilusky, Eric Martz, Alexander Berchansky, Joel L. Sussman, Eran Hodis, David Canner, Margaret Franzen