Human APP Intracellular Domain Complex with Fe65-PTB2
From Proteopedia
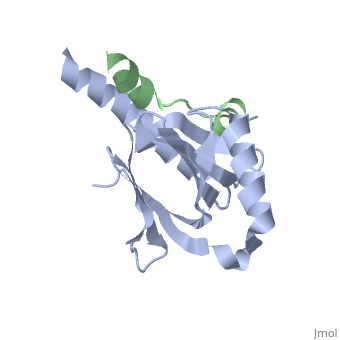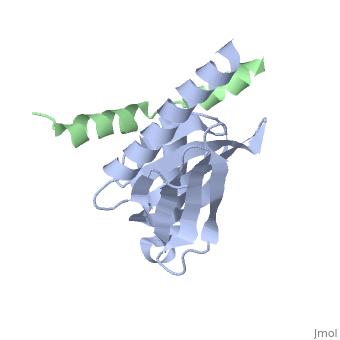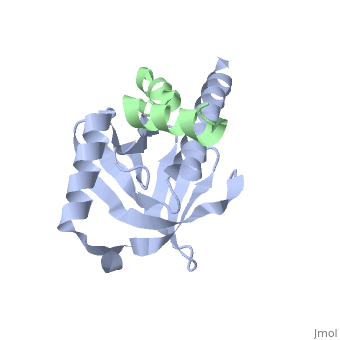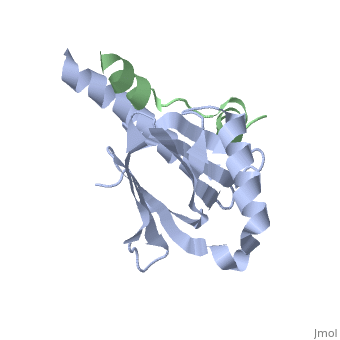
Intracellular domain of human APP in complex with Fe65-PTB2
Cleavage of the amyloid precursor protein (APP) is a crucial event in Alzheimer disease pathogenesis that creates the amyloid-beta peptide (Abeta) and liberates the carboxy-terminal APP intracellular domain (AICD) into the cytosol. [1] The study of interaction of the AICD and Fe65 protein is rather important, as Fe65 protein seems to be implied in production of Abeta and in signalling in APP.
Contents |
Structure
This structure contains 4 chains. . Each contains 140 residues: 4 helices and 7 strands. It's a part of the protein Fe65 binded with APP intracellular domain. . Each contains 35 residues: 2 helices and 1 strand. They represent APP intracellular domain.
The crystal structure of the APP intracellular domain is in complex with the . The interaction of the APP C terminus with the adaptor protein Fe65 mediates APP trafficking and signalling, and is thought to regulate APP processing and Abeta generation. The unique interface involves the NPxY PTB-binding motif and two alpha helices. The amino-terminal helix of the APP intracellular domain is , it's an Alzheimer disease-relevant phosphorylation site which is involved in Fe65-binding regulation. The structure together with mutational studies, isothermal titration calorimetry and nuclear magnetic resonance experiments sets the stage for understanding T(668) phosphorylation-dependent complex regulation at a molecular level.[2] Mutation at Thr-668 of APP abolished the effect of Fe65 on APP maturation. This mutation blocked the Fe65-dependent suppression of Abeta production and resulted in the release of increased levels of Abeta in the presence of Fe65 [3] .
Functions of human APP and Fe65 protein
There are suggestions that the binding of APP with Fe65 has been implicated in regulating cell motility and growth cone dynamics [4] [5]
Fe65 is an adaptor protein localized in the nucleus. It interacts with the Alzheimer's disease amyloid precursor protein (APP), transcription factor CP2/LSF/LBP1 and the low-density lipoprotein receptor-related protein. APP functions as a cytosolic anchoring site that can prevent the gene product's nuclear translocation. This encoded protein could play an important role in the pathogenesis of Alzheimer's disease. It is thought to regulate transcription. Also it is observed to block cell cycle progression by downregulating thymidylate synthase expression. Multiple alternatively spliced transcript variants have been described for this gene but some of their full length sequence is not known.[provided by RefSeq].[6]
Fe65 plays a central role in the response to DNA damage by translocating to the nucleus and inducing apoptosis. May act by specifically recognizing and binding histone H2AX phosphorylated on 'Tyr-142' (H2AXY142ph) at double-strand breaks (DSBs), recruiting other pro-apoptosis factors such as MAPK8/JNK1.Required for histone H4 acetylation at double-strand breaks (DSBs).Its ability to specifically bind modified histones and chromatin modifying enzymes such as KAT5/TIP60, probably explains its trancription activation activity. [7]
Location
Fe65 could be located in cell membrane, cytoplasm, nucleus. In normal conditions, it mainly localizes to the cytoplasm, while a small fraction is tethered to the cell membrane via its interaction with APP. Following exposure to DNA damaging agents, it is released from cell membrane and translocates to the nucleus.[8] APP is a cell surface protein that rapidly becomes internalized via clathrin-coated pits. During maturation, the immature APP (N-glycosylated in the endoplasmic reticulum) moves to the Golgi complex where complete maturation occurs (O-glycosylated and sulfated). After alpha-secretase cleavage, soluble APP is released into the extracellular space and the C-terminal is internalized to endosomes and lysosomes. Some APP accumulates in secretory transport vesicles leaving the late Golgi compartment and returns to the cell surface. [9]
APP and Alzheimer disease
Aβ peptides are generated in neuronal secretory vesicles by proteolytic cleavage of the amyloid precursor protein (APP) by proteases, called β-secretase and γ-secretase that cleave at the N-terminus and variant C-termini of Aβ within APP, respectively, resulting in Aβ of 40 or 42 amino acids (Aβ40 and Aβ42, respectively) [10]