Sandbox136
From Proteopedia
| |||||||||
| 1mg6, resolution 1.60Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Activity: | Phospholipase A(2), with EC number 3.1.1.4 | ||||||||
| |||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Contents |
Introduction
Anum-II is a Lys49 Phospholipase A2 (PLA2),protein extracted from Atropoides nummifer (a snake from central america). Lys49PLA2s are widespread in the venom of several vipers and are structurally classified as group II PLA2s, although the molecular mechanism of their myotoxicity remains unknown. K49-PLA2 possess the ability to disrupt biological membrane which lead to a significant muscle-tissue loss and disability in severely envenomed patients.
Structure
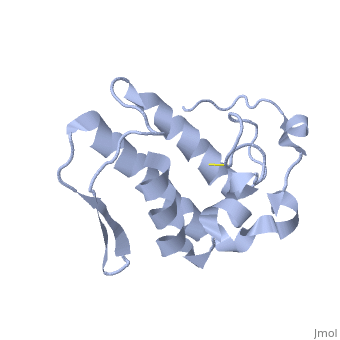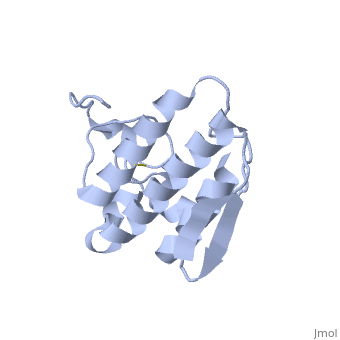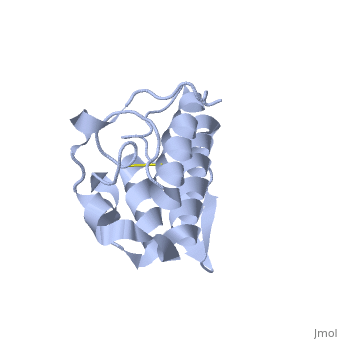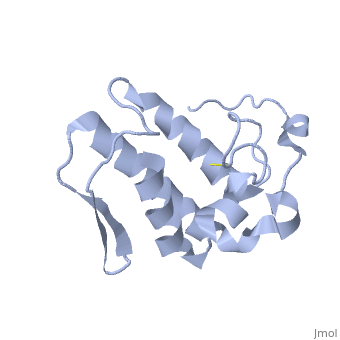
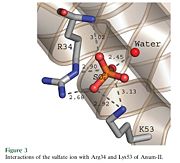
Anum-II has a length of 134 amino-acids. The phospholipase is formed by a short N-terminalαhelix (between residues 2-12), a 2nd αhelix (residues 40-55), two-stranded antiparallel linked thanks to a βwing (74-85) and a 3rd αhelix (residues 90-107). The 3rd αhelix is bound to the 2nd αhelix (in an antiparallele way) thanks to disulfure bonds ([Cys 44-Cys 105] and [Cys 51-Cys 98]) and thus form a . The protein is stabilized by 5 other disulfides bonds [Cys 27-Cys 125], [Cys 29-Cys 45] [Cys 50 Cys 134] [Cys 61-Cys 91] [Cys84-Cys96]. Alignment of Anum-II with other PLA2 have revealed that the positions of amino-acid residues which form the catalytic apparatus are conserved (His48,Tyr52, Tyr73 and Asp99) except for Asp49 which is replaced by Lys 49. The structure of the protein has revealed the presence of an anion-binding site between , and a water molecule. Sulfate ion is anchored thanks to hydrogen bonds between:
+O1 atom whit R34 Nε , K53 Nζ and a solvent water molecule.
+O2 atom whit the NH main chain of R34
+O4 atom whit K53 Nζ and R34 Nη2
This anion-binding site is supposed to play a role in the fixation of inhibitors but his function remains unknown. In the crystallization state (pH5), the protein is in a monomer state whereas in a solution at physiological pH, a dimerization occurs . The dimerization happened thanks to interactions at the interface of the N-terminal and β-wing regions . Studies have shown that Lys49 PLA2 is active only in the dimer state
Method of Determination
Venom was collected from more than 15 snakes and then purified by cation-exchange chromatography on carboxymethyl-Sephadex. Then single crystals were obtain whit the handing-drop vapour-diffusion method, and crystals were flash-cooled in crystallization solution with glycerol. After Structure was determined by X-Ray diffraction at 1,60 Å and a molecular replacement whit a specific program (MOLREP).
Physiological effect
Lys49 PLA2 has a strong myotoxicitic effect. The replacement of Asp49 by provides a particular role of the protein. Indeed, Asp49 allows the binding of Ca2+ an essential co-factor which is responsible for the activation of the catalytically apparatus (thanks to conserved amino-acids:His48, Tyr52, Tyr73 and Asp99). Lys49 PLA2 is not able to bind the co-factor, and thus is catalytically inactive. But some recent studies have shown that the Lys49 PLA2 is able to bind to the KDR (receptor of the VEGF165 growth factor)and this corresponds to his myotoxic activity.
Vascular endothelial growth factor (VEGF165) and its receptor Kinase insert domain containing receptor (KDR) are used in mechanisms of regulations of blood vessel formations. Experiments have revealed that the venom extracted from Agkistrodon piscivorus contains a KDR binding protein.It has been shown that this KDRb-p was in fact a Lys49 PLA2.Lys49 is able to bind to the extracellular domain of KDR receptors with a sub-nanomolar affinity. Recently studies revealed the fact that the region of the protein involved in the fixation on KDR was located in the C-terminal region. C- terminal region differ from the rest of the protein in his amino acid sequence, indeed some amino-acids are positive (K,R) and seem to play a role in the recognition and fixation on KDR.