NADH quinone oxidoreductase (NQO1) with inhibitor dicoumarol
From Proteopedia
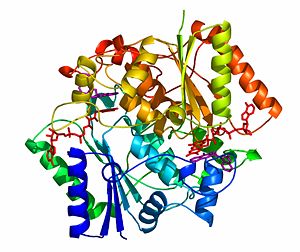
The crystal structure of NADH quinone oxidoreductase (NQO1) in complex with its potent inhibitor dicoumarol
NAD(P)H quinone oxidoreductase 1 (NQO1) is a ubiquitous flavoenzyme that catalyzes two electron reduction of quinones to hydroquinones utilizing NAD(P)H as an electron donor. NQO1 is a homo-dimer that functions via a “ping pong” mechanism. NAD(P)H binds to NQO1, reduces the FAD co-factor and is then released, allowing the quinone substrate to bind the enzyme and to be reduced. The NAD(P)H and the quinone binding sites of NQO1 have a significant overlap, thus providing a molecular basis for this “ping pong” mechanism.
Certain coumarins, flavones and the reactive dye cibacron blue are competitive inhibitors of NQO1 activity, which compete with NAD(P)H for binding to NQO1. Dicoumarol (3-3’–methylene-bis (4-hydroxycoumarin)), is the most potent competitive inhibitor of NQO1. Dicoumarol competes with NAD(P)H for binding to NQO1 and prevents the electron transfer to FAD.In addition to its role in the detoxification of quinones, NQO1 is also a 20S proteasome-associated protein that plays an important role in the stability of the tumor suppressor p53 and several other short-lived proteins including p73α and ornithine decarboxylase (ODC). NQO1 binds and stabilizes p53, protecting p53 from ubiquitin-independent 20S proteasomal degradation. Dicoumarol and several other inhibitors of NQO1 activity, which compete with NADH for binding to NQO1, disrupt the binding of NQO1 to p53 and induce ubiquitin-independent p53 degradation.
|
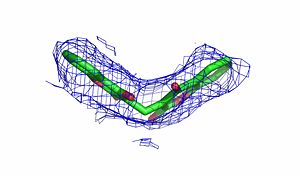
The crystal structure of human NQO1 in complex with dicoumarol was determined at 2.75 Å resolution (2f1o). NQO1 is a composed of two interlocked monomers. are formed and are present at the dimer interface (FAD is colored red and dicoumarol is colored blue). Therefore, each from these two is formed by both monomers. Dicoumarol is colored cyan, FAD in orange, nitrogens and oxygens are shown in CPK colors. NQO1 chain A is colored blueviolet and chain C in lime. NQO1 residues, participating in ligand interactions, are shown as stick representation and are labeled (A and C refer to the NQO1 chains). H-bonds are shown by dashed lines with their distances.
Structural comparison of the apo hNQO1 dimer (PDB accession code 1D4A in cyan) with hNQO1 in complex with dicoumarol(pink) reveals that structural changes associated with dicoumarol binding occur on several residues involving both monomers. The most prominent conformational changes that occur in the presence of dicoumarol involve Tyr 128 and Phe 232 that are present on the surface of the NQO1 catalytic pocket. Based on the comparison of NQO1 structure in complex with different NQO1 inhibitors and our previous analysis of NQO1 mutations that affect NQO1 interactions we propose that the specific conformation of Tyr 128 and Phe 232 is important for NQO1 interaction with p53 and other client proteins.
sna
Proteopedia Page Contributors and Editors (what is this?)
Alexander Berchansky, Orly Dym, Michal Harel, Jaime Prilusky, Moshe Ben-David, Joel L. Sussman, David Canner, Eric Martz