User:Liz Thomas/Sandbox 1
From Proteopedia
|
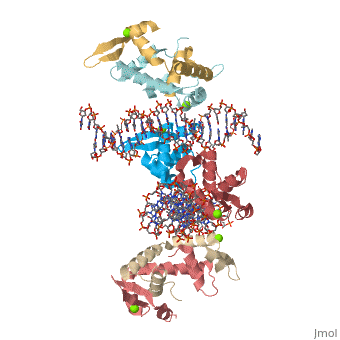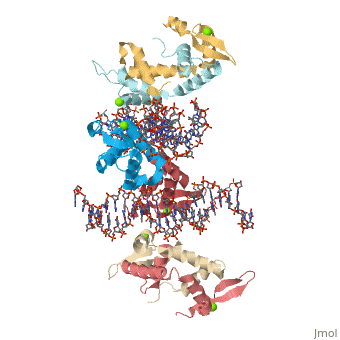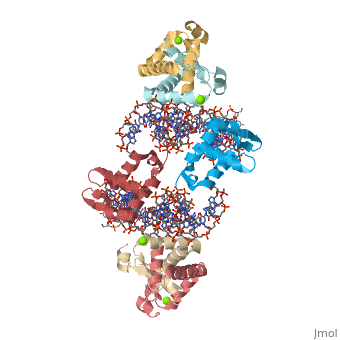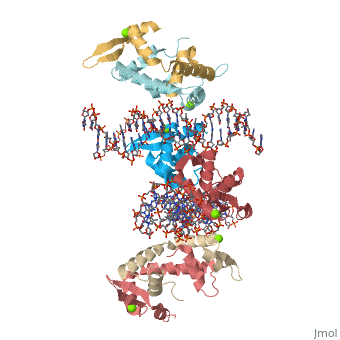
Crystal Structure of Foxp2 bound Specifically to DNA
New Article
|
| |||||||||
| 2a07, resolution 1.90Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | |||||||||
| Gene: | FOXP2, CAGH44, TNRC10 (Homo sapiens) | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Contents |
Introduction
Evolutionary Conservation
DNA Binding
Domain Swapping
Disease Mutations
Disease mutations in FOXP2 and related proteins correspond to either the domain-swapping dimer interface or the DNA binding sequence. FOXP2 and FOXP3 are similar enough that the crystal structure of one can be used to explain the effects of mutation on structure in the other.
Arg553His Mutation
This is the only mutation that has been characterized in the original FOXP2 protein.
Ile363Val Mutation
Ala385Thr Mutation
Arg397Trp Mutation
Phe371Cys and Phe371Leu Mutations