Pyrrolysyl-tRNA synthetase
From Proteopedia
| Please do NOT make changes to this Sandbox until after April 23, 2010. Sandboxes 151-200 are reserved until then for use by the Chemistry 307 class at UNBC taught by Prof. Andrea Gorrell. |
Contents |
Pyrrolysyl-tRNA synthetase
Introduction
Pyrrolysyl-tRNA synthetase(PyIRS)is encoded by the gene pyIS and belongs as a part of the group of enzymatic proteins whose role invovlves the cellular process of tRNA aminoacylation required for protein translation.[1] In particular, PyIRS is required for the activation of the amino acid pyrrolysine as it associates with a tRNA generating a specific tRNAPyl which is then further used to transfer the amino acid to a growing polypeptide.[2] The involvement of PyIRS is carried out due to the anticodon CUA on the supressor tRNAPyl that is complementary to the UAG codon.[3][4] The interesting fact is that this is done by the response of the codon UAG (amber codon) on the mRNA that is normally a stop codon in other organisms. Pyrrolysine (Pyl) is the 22nd existing amino acid genetically encoded in nature that was first discovered as a byproduct contained by the active site of monomethylamine methyltransferase exclusively from Methanosarcina barkeri (M. barkeri) species.[2][5] Thus, it is utalized by a variety of organisms that metabolize methylamines for aquiring energy such as methanogenic Archaea of the family Methanosarcinace; along with two known bacterium species[1][5] Pyrrolysine’s structural makeup consists of 4-methylpyrroline-5-carboxylate in amide linkage with the ϵN of lysine.[6] This arrangement is comparable to lysine; however; however, being its derivative it contains an added pyrroline ring situated at the back of the structure.[6]
In addition, by observing M. barkeri cellular mechanisms containing PyIRS it was detected that it furthermore has the capability to activate an assortment of other non-natural pyrrolysine/lysine derivatives; as well as the non-canonical amino acids.[7] These amino acids can then be further added to their specialized tRNAPyl; resulting in them forming new polypeptides. This procedure is performed my extracting PyIRS, and the amber suppressor tRNAPyl from Methanosarcina. When obtained a scientifically modified tRNA/aminoacyl-tRNA synthetase (aaRS) pair must be designed for the recognition and for the unique aminoacylation intended for the selected amino acid.[3] Once this has been completed it then is carefully placed into a bacterium species such as Escherichia coli (E. coli).[3] The reason that this is successful is because once inserted tRNAPyl function as a orthogonal pair with aaRS-tRNA that will not interfere with cellualr mechanisms and other components of translation.[2][3] In particular, Nϵ-(tert-butyloxycarbonyl –L-lysine (BocLys)is a non-natural amino acid that is a derivative of lysine which can be intergraded into polypeptides utilizing the amber codon by the process of being esterified to tRNAPyl by PyIRS in E.coli for the incorporation into proteins.[2] By using this method we can obtain proteins with manipulated structures and functions that can serve useful purposes in studying cellular processes and in altering further mechanisms.
Structure
|
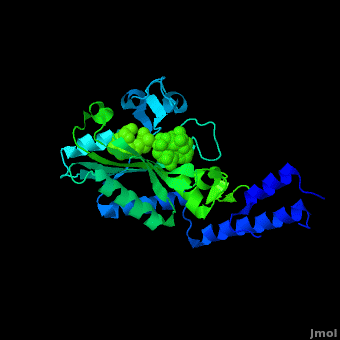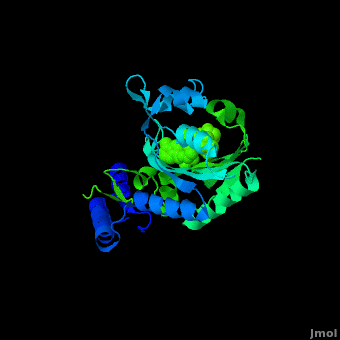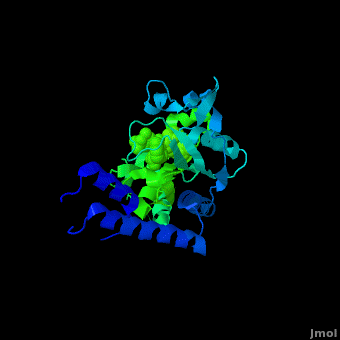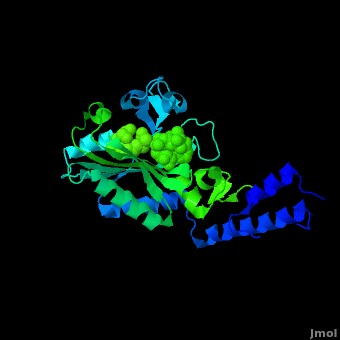
Pyrrolysyl-tRNA synthetase (PyIRS) attached with ; along with adenosine 5’ (beta, gamma-imido) triphosphate (AMMPPNP)demonstrates the proposed structure that is required for the efficient recognition of amino acids and the aminoacylation by PyIRS.[2] As shown above it consists of a multidomain polypeptide made of 1 chain (Chain A) comprising of a total length of 291 residues, consisting of 9 (95 residues) making 32% of the structure, and the remaining is 12 (59 residues) consisting of 20% of the other structure. In addition, its structure includes 4 ligands; ANP, EDO, LBY, and MG, as well as two catalytic domains; PRK06253 and class_II_aaRS-like_core that is used for the fermentation of the enzyme bound aminoacyl-adenylate in the presence of ATP. This complex was determined at 1.79 Anstroms and with the visible electon density in the active site it was experimentally trialed that the Ne-Boc group is situated in the hydrophobic interior similarly to the standard pyrrolysine AMPPNP bound arrangement.[2] Having the Ne-BocLys positioned in this way it has the capability to hydrogen bond with the amide group side chain of Asn346.[2] Next, the Cα-carbonyl groups of BocLys in turn will hydrogen bond Asn346 contrary to the α-amino group which is linked to α-phosphate group of AMPPNP.[2] In order for the substrate to effectively bind to the side chain amide group Asn354 will inducible fit the carbonyl group of the substrate pyrrolysine and BocLys into position.[2] Additionally, the BocLys α-carboxyl group is directed to that it is associated outside from the active site allowing for the flexibility due to the ability to rotate around the Cɑ- Cβ bond.[2]
Mechanism
Th proposed mechasim for the insertion of pyrrolysine into polypeptides initially begins with the activation of a special tRNA. This is completed by the charging of tRNA as it interacts with lysine via aminoacylation by PyIRS in the presence of the exchange of ATM for AMP and PPi (inorganic pyrophosphate). This will generate lysyl-tRNACUA which is then pre-translationally modified by the influence of PyIB, PyIC and PyID genes forming Pyl-tRNACU. This specific tRNAPyl is used to transfer pyrrolysine to the A-site in the ribisome which is then added the co- translated growing polypeptide chain. To ensure the appropriate the amino acids find its way to PyIRS, and not to any other class II aaRS present, PyIRS has special identification mechanisms associated it. These include specific characteristics such as its size, bulkiness according how it is going to bind to the hydrophobic position.[2] In addition, appropriate hydrogen bond acceptors/donors must be positioned adjacent Asn346, and the appropriate length of the side-chain spacer must be available to make certain the binding is excellent for the reaction to proceed efficiently.[2] For the proper recognition of the appropriate amino acid there must be Ne-carbonyl group that has a specific size for the substituent.[2]
References
- ↑ 1.0 1.1 Herring S, Ambrogelly A, Polycarpo CR, Soll D. Recognition of pyrrolysine tRNA by the Desulfitobacterium hafniense pyrrolysyl-tRNA synthetase. Nucleic Acids Res. 2007;35(4):1270-8. Epub 2007 Jan 31. PMID:17267409 doi:10.1093/nar/gkl1151
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 Yanagisawa T, Ishii R, Fukunaga R, Kobayashi T, Sakamoto K, Yokoyama S. Multistep engineering of pyrrolysyl-tRNA synthetase to genetically encode N(epsilon)-(o-azidobenzyloxycarbonyl) lysine for site-specific protein modification. Chem Biol. 2008 Nov 24;15(11):1187-97. PMID:19022179 doi:10.1016/j.chembiol.2008.10.004
- ↑ 3.0 3.1 3.2 3.3 Urbancsek J, Rabe T, Grunwald K, Kiesel L, Papp Z, Runnebaum B. High preovulatory serum luteinizing hormone level is unfavorable to conception. Gynecol Endocrinol. 1991 Dec;5(4):223-33. PMID:1796745
- ↑ Polycarpo C, Ambrogelly A, Berube A, Winbush SM, McCloskey JA, Crain PF, Wood JL, Soll D. An aminoacyl-tRNA synthetase that specifically activates pyrrolysine. Proc Natl Acad Sci U S A. 2004 Aug 24;101(34):12450-4. Epub 2004 Aug 16. PMID:15314242 doi:10.1073/pnas.0405362101
- ↑ 5.0 5.1 Nozawa K, O'Donoghue P, Gundllapalli S, Araiso Y, Ishitani R, Umehara T, Soll D, Nureki O. Pyrrolysyl-tRNA synthetase-tRNA(Pyl) structure reveals the molecular basis of orthogonality. Nature. 2009 Feb 26;457(7233):1163-7. Epub 2008 Dec 31. PMID:19118381 doi:10.1038/nature07611
- ↑ 6.0 6.1 Soares JA, Zhang L, Pitsch RL, Kleinholz NM, Jones RB, Wolff JJ, Amster J, Green-Church KB, Krzycki JA. The residue mass of L-pyrrolysine in three distinct methylamine methyltransferases. J Biol Chem. 2005 Nov 4;280(44):36962-9. Epub 2005 Aug 11. PMID:16096277 doi:10.1074/jbc.M506402200
- ↑ Polycarpo CR, Herring S, Berube A, Wood JL, Soll D, Ambrogelly A. Pyrrolysine analogues as substrates for pyrrolysyl-tRNA synthetase. FEBS Lett. 2006 Dec 11;580(28-29):6695-700. Epub 2006 Nov 20. PMID:17126325 doi:10.1016/j.febslet.2006.11.028
Proteopedia Page Contributors and Editors (what is this?)
Agatka Jaworski, Michal Harel, Joel L. Sussman, David Canner, Andrea Gorrell, Alexander Berchansky