Apurinic-Apyrimidinic Endonuclease
From Proteopedia
Contents |
APURINIC/APYRIMIDINIC ENDONUCLEASE-1 (APE1)
INTRODUCTION
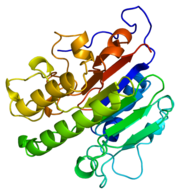
DNA is damaged frequently in living cells via endogenous and exogenous factors that have the ability to cause abasic sites in the DNA strand. Abasic DNA must be repaired both accuratly and efficiently in order to prevent the death of the cell or the accumulation of mutations within the genome. Abasic DNA is repaired in the base excision repair pathway. Apurinic/apyrimidinic endonuclease-1 (APE1) is the major repair enzyme in the base excision repair pathway in humans [2]. APE1 has the ability to cleave the phophodiester bond at the 5' end of abasic sites in damaged DNA strands [3]. An important point of gene regulation within a cell is the regulation of the half life of mRNA transcripts. The regulation of mRNA transcripts will control the amount of protein translated in a cell. Many mRNA transcripts have been linked to diseases such as cancer. These deleterious mRNA transcripts are known as proto-oncogene products and can greatly effect the health of cells[4]. APE1 has also been shown to have the ability to regulate the half life of c-myc mRNA by endonucleolytically cleaving the coding region determinant (CRD) of c-myc mRNA[5]. The oxidation states of molecules, such as many transcription factors, within the cell can activate or deactivate molecules and therefore regulate the expression of genes [6]. The oxidation states of many amino acid residues within a protein can change the properties of the protein which can greatly affect the enzymatic activity of the protein. The oxidation states of cysteine residues, among other amino acid residues, within a polypeptide are regulated by APE1[7]. Proteins that APE1 has been established to regulate the oxidation states of the cysteine residues of include NF-kB, Ehr-1, HIF-1, HLF, Pax-5, Pax-8 and p53, the mechanism of how APE1 contributes to the oxidation states of the cysteine residues on these proteins, however, is poorly understood[8]. It is due to these many functions of APE1 that APE1 has become known as the multifunctional workhorse protein of the cell.
STRUCTURE AND FUNCTION
|
APE1 is classified as a DNA lyase that has a structure weight of 50000-75000Da, depending on the crystal structure in question [9]. Many crystal structures of APE1 have been determined with amino acid numbers ranging from ~270 amino acid residues to ~320 amino acid residues[10][11].
Substrate Binding and Enzymatic Mechanism
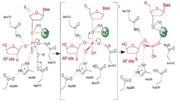
APE1 has numerous substrates in which in binds to ands acts on enzymatically with abasic DNA being the most well known substrate for APE1. APE1 binds on the 5' side of the abasic DNA site with the amino acid residues Arg73, Ala74 and Lys78 interacting with three consecutive phosphate molecules on the DNA strand, while Tyr128 and Gly127 simultaneously span the minor grove causing the groove to widen a distance of ~2 Angstroms[12]. The DNA is anchored and the abasic DNA is stabalized by APE1. APE1 then inserts Met270 into the minor groove causing the APE1/DNA structure to become extremely kinked and this kinking of the DNA strand is followed by an Arg177 insertion to the major groove donating a hydrogen bond to the 3' phosphate located at the abasic site[13]. This is followed by a nucleophilic attack of an aspartate-activated hydroxyl group forming a transition state that is stabalized by a Mg^2+ ion which is followed by cleavage of the phosphodiester bond on the DNA strand[14]. This is followed by the sequential events of residue Trp280 contacting a DNA phosphate and residues Asn222, Asn226 and Asn229 contacting the phosphates 3' of the abasic site, which is followed by Asn174 and Asn212 forming hydrogen bonds with an oxygen molecules 5' to the abasic site[15].
The nuclease domain
The redox domain
APE1 has been established as a vital redox enzyme that plays pivotal roles in repairing both damaged DNA and reducing transcription factors to promote higher binding affinity with DNA. Although the central dogma for mRNA decay currently favours the 3’ to 5’ deadenylation-dependent and the 5’ to 3’ decapping decay pathways, endonucleolytic cleavage intermediates have been shown to be involved in the decay of a number of mRNA (Barnes 2009). Many mRNAs, such as c-myc mRNA, have been shown to be degraded by 3’-5’ exonucleolytic activity via exonucleases (Barnes 2009). In the paper Identification of Apurinic/apyrimidinic endoribonuclease 1 (APE1) as the endoribonuclease that cleaves c-myc mRNA in vitro, Barnes et al. show that the endonuclease APE1/Ref1 is responsible for the endonucleolytic cleavage of c-myc mRNA in vitro. The vital role of APE1 now includes endonucleolytic cleavage of c-myc mRNA along with its role in redox repair of damaged DNA bases.
POPULATION VARIENTS
APE1 HAS BEEN SHOWN TO CLEAVE C-MYC MRNA
REFERENCES
- ↑ 1e9n
- ↑ Masood, Z.H. 2000. Functional characterization of APE1 varients identified in the human population. Nucleic Acid Research 28: 3871-3879.
- ↑ Masood, Z.H. 2000. Functional characterization of APE1 varients identified in the human population. Nucleic Acid Research 28: 3871-3879.
- ↑ Barnes, T., et al. 2009. Identification of Apurinic/apyrimidinic endoribonuclease 1 (APE1) as the endoribonuclease that cleaves c-myc mRNA in vitro. Nucleic Acid Research 37: 3946-3958.
- ↑ Barnes, T., et al. 2009. Identification of Apurinic/apyrimidinic endoribonuclease 1 (APE1) as the endoribonuclease that cleaves c-myc mRNA in vitro. Nucleic Acid Research 37: 3946-3958.
- ↑ Ando, K. et al. 2008. A new APE1/Ref-1-dependent pathway leading to reduction of NF-kB and AP-1, and activation of their DNA binding activity. Nucleic Acid research 36(13): 4327-4336.
- ↑ Ando, K. et al. 2008. A new APE1/Ref-1-dependent pathway leading to reduction of NF-kB and AP-1, and activation of their DNA binding activity. Nucleic Acid research 36(13): 4327-4336.
- ↑ Ando, K. et al. 2008. A new APE1/Ref-1-dependent pathway leading to reduction of NF-kB and AP-1, and activation of their DNA binding activity. Nucleic Acid research 36(13): 4327-4336.
- ↑ Mol,C.D. et al. 2000. DNA-bound structures and mutants reveal abasic DNA binding by APE1 DNA repair and coordination. Nature 403:451-456.
- ↑ Mol,C.D. et al. 2000. DNA-bound structures and mutants reveal abasic DNA binding by APE1 DNA repair and coordination. Nature 403:451-456.
- ↑ Masood, Z.H. 2000. Functional characterization of APE1 varients identified in the human population. Nucleic Acid Research 28: 3871-3879.
- ↑ Mol,C.D. et al. 2000. DNA-bound structures and mutants reveal abasic DNA binding by APE1 DNA repair and coordination. Nature 403:451-456.
- ↑ Mol,C.D. et al. 2000. DNA-bound structures and mutants reveal abasic DNA binding by APE1 DNA repair and coordination. Nature 403:451-456.
- ↑ Mol,C.D. et al. 2000. DNA-bound structures and mutants reveal abasic DNA binding by APE1 DNA repair and coordination. Nature 403:451-456.
- ↑ Mol,C.D. et al. 2000. DNA-bound structures and mutants reveal abasic DNA binding by APE1 DNA repair and coordination. Nature 403:451-456.
- ↑ 1e9n
Proteopedia Page Contributors and Editors (what is this?)
Mark Barnes, Michal Harel, Alexander Berchansky, David Canner, Jaime Prilusky, Andrea Gorrell, Eran Hodis, Joel L. Sussman
