Sandbox 45
From Proteopedia
Structure of Trypsin
The various interactive tendencies and chemical characteristics of amino acids in this serine protease contribute to the protein's structure and catalytic function. The spacial arrangement of Trypsin's 223 residues in relation to themselves and their aqueous environment is displayed below.
Secondary Structure
Bovine Trypsin contains three of lengths XYandZ. The two , A and B, are comprised of 7 and 6 strands. Although both appear as such, only B is technically a beta barrel. In the native conformation, these regular secondary structures interact with themselves and one another at a number of locations by numerous forces of attraction. A closer look at helix X,terminal, shows , between the helix and local residues of the remaining peptide. This is significant to its role in the 3D structure of the protein.
|
The of hydrophilic (green) and hydrophobic (yellow) residues is one of the most important aspects of primary structure. The protein as a whole achieves its native conformation primarily by the hydrophobic collapse of supersecondary structure; hydrophobic side chains are internalized while water molecules interact with the water-soluble side chains pushed to the exterior. The water's (red) with the surface of the protein shows this, as a view shows an absence of water within the hydrophobic core.
also contribute to the stability of the protein. Typically, proteins in an extra-cellular, oxidizing environment contain disulfide bonds that hold the structure together through variable temperature and pH. It follows that trypsin, a digestive protease found in the digestive tract, would require this added stability.
Ligand Binding and Catalysis
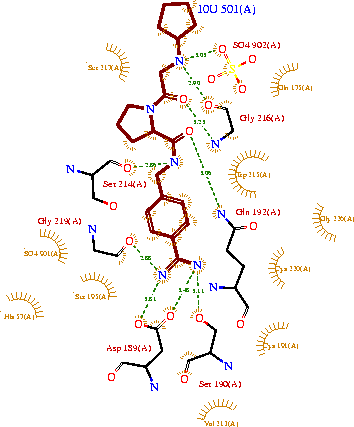
The structure of this particular bovine trypsin was determined in complex with , formula C20H29N5O2, along with two (highlighted) and a Calcium ion (green). Four key amino acids interact with Calcium at a . The binding of ligand 10U involves , direct , and a host of . The figure below shows this binding in two dimensions.
The binding of trypsin to ligand 10U somewhat emulates the binding to its specific peptide substrates. The preference for lysine or arginine in trypsin catalysis is due to the side chains of the trypsin . Here (green), aspartic acid 189 and one of two significant glycine backbones, Gly 216, interact with the ligand as they would with Arg or Lys.
The ; Asp 102, His 57, and Ser 195, shown here in yellow, is positioned near the substrate. The catalytically active histidine and serine side chains are even near an amide bond in 10U, just like the amide bond broken in peptide hydrolysis. According to FirstGlance in Jmol, there is no bonding of these groups with the ligand, apart from minor hydrophobic interactions with Hist 57. If Ligand 10U were a transition state analog, some covalent connection would exist in addition to hydrogen bonds. 10U simulates the substrate, but does not hydrolyze at either of its two amide bonds, likely due to local groups atypical of peptide backbones.