Sandbox 48
From Proteopedia
Template:Tims Sandbox Reservation
|
General Preliminary Information and Historical Development
Lysozyme is an enzyme known for its unique ability to degrade the polysaccharide architecture of many kinds of cell walls, normally for the purpose of protection against bacterial infection[1]. In avian species, lysozyme is an abundant component of egg white, in which lysozyme functions as an antibiotic and as a nutrient for early embryogenesis [2]. In vertebrate species, lysozyme is generally found in mucosal secretions, such as tears and saliva, and functions in the same way that it functions in avian species for antibacterial purposes.
The discovery of lysozyme in 1922 by Alexander Fleming was providential in that the undertaken experiment related to the discovery of lysozyme was not geared toward any knowledge of such a protein as lysozyme [3]. During the unrelated experiment, nasal drippings were inadvertently introduced to a petri dish containing a bacterial culture, which culture consequently exhibited the results of an as yet unknown enzymatic reaction. The observation of this unknown reaction led to further research on the components of this reaction as well as to the corresponding identification of the newfound "lysozyme." Fleming's discovery was complemented by David C. Phillips' 1965 description of the three-dimensional structure of lysozyme via a 200pm resolution model obtained from X-ray crystallography [4]. Phillips' work was especially groundbreaking since, by successfully elucidating the structure of lysozyme via X-ray crystallography, Phillips had managed to successfully elucidate the structure of an enzyme via X-ray crystallography - a feat that had never before been accomplished[5]. Phillips' research also led to the first sufficiently described enzymatic mechanism of catalytic action [6]. Thus, Phillips' elucidation of the function of lysozyme led Phillips to reach a more general conclusion on the diversity of enzymatic chemical action in relation to enzymatic structure. Clearly, the historical development of the understanding of the structure and function of lysozyme has been paramount to the more general realm of enzyme chemistry.
General Function
Lysozyme is a 129 amino acid-long enzyme that is particularly specific in its cleavage proclivity for alternating polysaccharide copolymers of N-acetyl glucosamine (NAG) and N-acetyl muramic acid (NAM), which architectural theme represents the "unit" polysaccharide structure of many bacterial cell walls [7]. The location of cleavage for lysozyme on this architectural theme is the β(1-4) glycosidic linkage connecting connecting the C1 carbon of NAM to the C4 carbon of NAG.
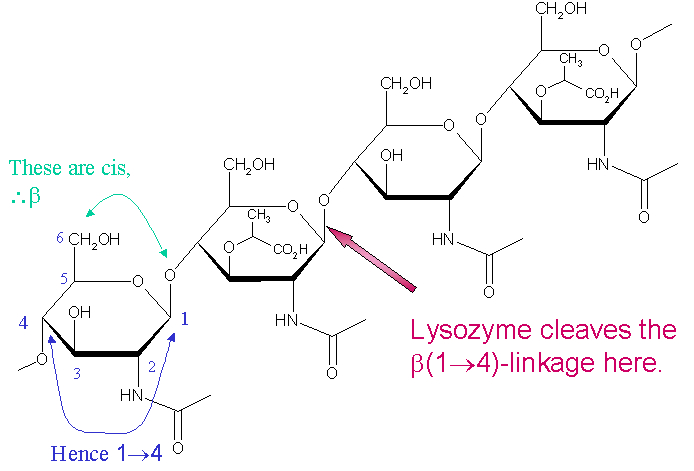 [8]
[8]
The particular substrate of preference for this cleavage type is a (NAG-NAM)₃ hexasaccharide, within which substrate occurs the cleaving target glycosidic bond of NAM₄-β-O-NAG₅. The individual hexasaccharide binding units are designated A-F, with the NAM₄-β-O-NAG₅ glycosidic bond cleavage preference corresponding to a D-E unit glycosidic bond cleavage preference.
Mechanism of Action
The lysozyme mechanism of action results in the hydrolysis of a glycoside, which corresponds to the conversion of an acetal to a hemiacetal, which reaction (general degradation of glycosidic bond to units "capped" by newly formed hydroxyl groups) necessitates acid catalysis, since the conversion of acetal to hemiacetal involves the protonation of the reactant oxygen prior to actual bond cleavage. [9]. Furthermore, the transition state obtained from this protonation is a covalent, oxonium ion, intermediate that must obtain resonance stabilization. The need for some means of acid catalysis and covalent resonance stabilization is adequately provided by the Glu 35 and Asp 52 residues of lysozyme, respectively. The reaction mechanism of lysozyme is demonstrated. In the following image, the reaction begins at the upper left-hand side, and proceeds according to reaction arrows.
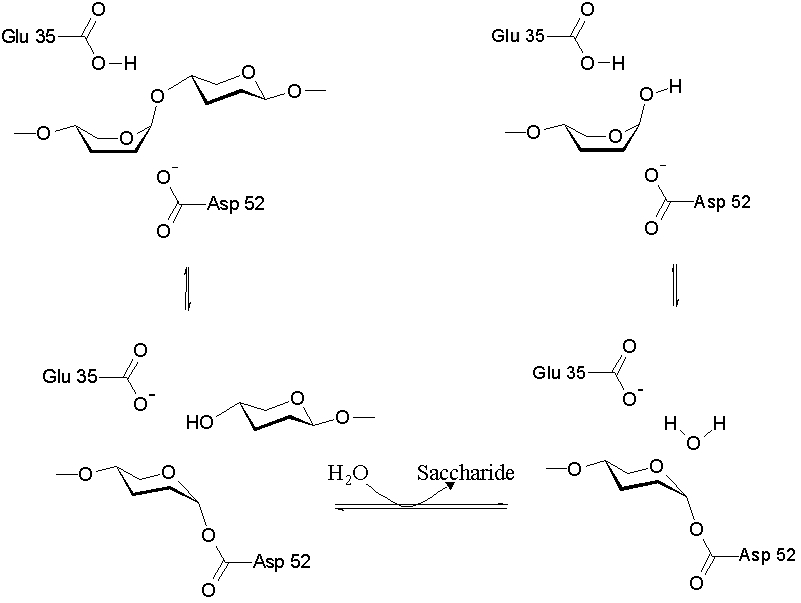 [10]
[10]
The active site of lysozyme is formulated as a prominent cleft outlined by the two aforementioned catalytic enzymes, Glu 35 and Asp 52.
- ↑ Lysozyme. 2010. Citizendium.org. http://en.citizendium.org/wiki/Lysozyme
- ↑ Sears, D.W. 2010. Overview of the Structure and Function of Hen Egg-White Lysozyme. Ucsb.edu. http://mcdb-webarchive.mcdb.ucsb.edu/sears/biochemistry/tw-enz/lysozyme/HEWL/lysozyme-overview.htm/
- ↑ Lysozyme. 2008. Lysozyme.co.uk. http://lysozyme.co.uk/
- ↑ Lysozyme, 2008. Lysozyme.co.uk. http://lysozyme.co.uk/
- ↑ Bugg, T. 1997. An Introduction to Enzyme and Coenzyme Chemistry. Blackwell Science Ltd., Oxford
- ↑ 1967. Proc R Soc Lond B Bio 167 (1009): 389–401.
- ↑ Sears, D.W. 2010. Overview of the Structure and Function of Hen Egg-White Lysozyme. Ucsb.edu. http://mcdb-webarchive.mcdb.ucsb.edu/sears/biochemistry/tw-enz/lysozyme/HEWL/lysozyme-overview.htm/
- ↑ Image from: http://www.vuw.ac.nz/staff/paul_teesdale-spittle/essentials/chapter-6/proteins/lysozyme.htm
- ↑ Pratt, C.W., Voet, D., Voet, J.G. Fundamentals of Biochemistry - Life at the Molecular Level - Third Edition. Voet, Voet and Pratt, 2008.
- ↑ Image from: http://www.google.com/imgres?imgurl=http://www.vuw.ac.nz/staff/paul_teesdale-spittle/essentials/chapter-6/pics-and-strucs/lysozyme-mech.gif&imgrefurl=http://www.vuw.ac.nz/staff/paul_teesdale-spittle/essentials/chapter-6/proteins/lysozyme.htm&usg=__ormapG4XKg-tR5GrMSOdSMTV4vE=&h=603&w=801&sz=7&hl=en&start=17&zoom=1&tbnid=nvr9gvFrUILDkM:&tbnh=143&tbnw=189&prev=/images%3Fq%3DThe%2Blysozyme%2Breaction%2Bmechanism%26um%3D1%26hl%3Den%26sa%3DN%26biw%3D1280%26bih%3D647%26tbs%3Disch:10%2C304&um=1&itbs=1&iact=hc&vpx=521&vpy=349&dur=448&hovh=191&hovw=254&tx=140&ty=48&ei=JQ_LTPKzLIjCsAPkzt2KDg&oei=IA_LTP74OsG78gapm-GFAQ&esq=2&page=2&ndsp=18&ved=1t:429,r:2,s:17&biw=1280&bih=647
