Phosphoinositide 3-Kinases
From Proteopedia
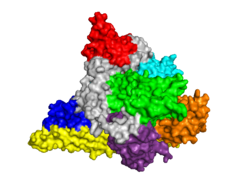
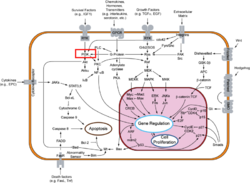
Template:STRUCTURE 3hhm Phosphoinositide 3-Kinases (PI3K) are a family of ubiquitously distributed lipid kinases, that play a critical role in the regulation of numerous cellular processes including cellular growth and morphology, programmed cell death, cell motility and adhesion, mitogenesis and glucose uptake. [1] PI3K generates important second messengers by catalyzing the transfer of the γ-phosphate group of ATP to the D3 position of phosphoinositides. [2] The PI3K preferred substrate is Phosphatidylinositol-4,5-bisphosphate (PIP2), which is converted into phosphatidylinositol-3,4,5-triphosphate (PIP3) upon phosphorylation at the cell membrane. The importance of PI3K is evident in knockout mice studies in which those mice with disruptions of critical PI3K components have significant deficiencies in immune and inflammatory response [3] sometimes resulting in embryonic death.[4] Aberrations in PIP3 levels, either through activation of PI3ks or through inactivation of lipid phosphatase PTEN, occur frequently in numerous forms of cancer, making PI3K an exciting new target to treat cancer among other human diseases.[5]
The Classes of PI3Ks
PI3Ks can be grouped into three distinct classes, Class I-III. Class I PI3Ks, the most well understood and thoroughly explored PI3K class, are composed of a 110kDa and a 50-100 kDa . Activation of Class I PI3Ks is controlled by extracellular signaling via receptors with intrinsic tyrosine kinase activity, G protein-linked receptors, or receptors coupled to SRC like protein tyrosine kinases. [6] Class II PI3Ks are relatively poorly understood but are 170-210 kDa and have in vitro substrate specificity toward PtdIns 4-P. Class III PI3Ks depend on Vps15p protein Ser/Thr kinases, which recruits the phosphatidylinositol kinase to late Golgi Compartments. [2]Class I Subclasses
PI3Ks are activated by extracellular agonists via the translocation of PI3Ks to the plasma membrane for easy access to lipid substrates. Depending on the adaptor proteins involved in the process, Class I PI3Ks are segregated into two subgroups. Those that associate with p85 will be directed to phosphorylated tyrosine motifs (Class IA), while PI3Kγ interacts with trimeric G proteins and the p101 protein (Class IB) [2]
Structure of PI3K
For Full Article, See: The Structure of PI3K
Class I PI3Ks, which are tightly regulated by tyrosine kinases, are composed of an 85kDa regulatory/adapter subunit (p85) and a 110kDa catalytic subunit (p110). [7]
PI3K Activation, Inhibition, and Medical Implications
For Full Article, See: PI3K Activation, Inhibition, & Medical Implications
A number of inhibitors for PI3K have been developed to understand how PI3K is activated and functions. These analysis have massive medical implications for the treatment of Cancer and Diabetes.
Additional 3D Structures
Solved Structures of PI3K
Class I PI3K
PI3K SH2 Domain
2iug, 2iuh, 2iui – Crystal Structure of PI3K nSH2 Domain with Peptides
1h9o – Crystal Structure of PI3K SH2 Domain with PDGFR Peptide
1fu5, 1fu6 – NMR structure of nSH2 Domain from PI3K
PI3K ISH2 Domain
3mtt – Crystal Structure of PI3K ISH2 Beta Crystal
3l4q – Crystal Structure of PI3K ISH2 in Influenza
2v1y – Crystal Structure of ISH2 in complex with ADB
PI3K SH3 Domain
3i5s, 3i5r – Crystal Structure of SH3 Domain in complex with peptide
2kt1 – Crystal Structure of SH3 Domain in p85 beta
1pht – Crystal Structure of PI3K Alpha SH3 Domain
1pks, 1pkt – Crystal Structure of PI3K SH3 Domain
p110 Subunit of PI3K
3lj3 – Crystal Structure of PI3K Gamma bound to Pyrrolopyridine-Benzofuran Inhibitor
3l54 – Crystal Structure of PI3K Gamma
3l13, 3l16, 3l17 – Crystal Structure of Pan-PI3-Kinase with Inhibitor
3l08 – Crystal Structure of PI3K Gamma bound to GSK2126458
3ibe – Crystal Structure of PI3K Gamma bound to Pyrazolopyrimidine Inhibitor
3hhm, 3hiz – Crystal Structure of p110 & NISH2
3ene – Crystal Structure of PI3K Gamma with inhibitor
3dpd – Crystal Structure of PI3K with oxazines inhibitor
3dbs – Crystal Structure of PI3K Gamma bound to GDC0941
3csf, 3cst – Crystal Structure of p110 Gamma bound to organourethenium inhibitor
2x38 – Crystal Structure of p110 Delta bound to IC87114
2wxf, 2wxg, 2wxh, 2wxi, 2wxj, 2wxk, 2wxl, 2wxm, 2wxn, 2wxo, 2wxp, 2wxq, 2wxr – Crystal Structure of p110 Delta with Inhibitors
2v4l – Crystal Structure of PI3K p110 Gamma with inhibitor
2rd0 – Crystal Structure of PI3K p110/p85 complex
2chw, 2chx, 2chz – Crystal Structure of PI3K Gamma with PIK-39 Inhibitor
2a4z, 2a5u – Crystal Structure of PI3K gamma complex with AS604850 and AS605240 Inhibitors
1he8 – Crystal Structure of RAS – PI3K Gamma Complex
1e7u, 1e7v, 1e7w, 1e7y, 1e7z, 1e90, 1e8x – Crystal Structure of PI3K Bound to Various Inhibitors
PI3K C2 Domain
2wwe – Crystal Structure of PI3K C2 Gamma Domain
2enq – Crystal Structure of C2 Domain, p110 Alpha
Class III PI3K
2x6f, 2x6h, 2x6j, 2x6k – Crystal Structure of Class III PI3K bound to various inhibitors
3ls8 – Crystal Structure of Class III PI3K in complex with inhibitor
3ihy – Crystal Structure of Human PI3K Class III
Additional Resources
- See: Cancer For Additional Proteins involved in the disease.
- See: Oncogenes for Additional examples of oncogenes and tumor suppressor genes.
References
- ↑ Djordjevic S, Driscoll PC. Structural insight into substrate specificity and regulatory mechanisms of phosphoinositide 3-kinases. Trends Biochem Sci. 2002 Aug;27(8):426-32. PMID:12151228
- ↑ 2.0 2.1 2.2 Wymann MP, Pirola L. Structure and function of phosphoinositide 3-kinases. Biochim Biophys Acta. 1998 Dec 8;1436(1-2):127-50. PMID:9838078
- ↑ Sasaki T, Irie-Sasaki J, Horie Y, Bachmaier K, Fata JE, Li M, Suzuki A, Bouchard D, Ho A, Redston M, Gallinger S, Khokha R, Mak TW, Hawkins PT, Stephens L, Scherer SW, Tsao M, Penninger JM. Colorectal carcinomas in mice lacking the catalytic subunit of PI(3)Kgamma. Nature. 2000 Aug 24;406(6798):897-902. PMID:10972292 doi:10.1038/35022585
- ↑ Bi L, Okabe I, Bernard DJ, Wynshaw-Boris A, Nussbaum RL. Proliferative defect and embryonic lethality in mice homozygous for a deletion in the p110alpha subunit of phosphoinositide 3-kinase. J Biol Chem. 1999 Apr 16;274(16):10963-8. PMID:10196176
- ↑ Miled N, Yan Y, Hon WC, Perisic O, Zvelebil M, Inbar Y, Schneidman-Duhovny D, Wolfson HJ, Backer JM, Williams RL. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science. 2007 Jul 13;317(5835):239-42. PMID:17626883 doi:317/5835/239
- ↑ Stephens LR, Hughes KT, Irvine RF. Pathway of phosphatidylinositol(3,4,5)-trisphosphate synthesis in activated neutrophils. Nature. 1991 May 2;351(6321):33-9. PMID:1851250 doi:http://dx.doi.org/10.1038/351033a0
- ↑ Hoedemaeker FJ, Siegal G, Roe SM, Driscoll PC, Abrahams JP. Crystal structure of the C-terminal SH2 domain of the p85alpha regulatory subunit of phosphoinositide 3-kinase: an SH2 domain mimicking its own substrate. J Mol Biol. 1999 Oct 1;292(4):763-70. PMID:10525402 doi:http://dx.doi.org/10.1006/jmbi.1999.3111
Proteopedia Page Contributors and Editors (what is this?)
David Canner, Michal Harel, Joel L. Sussman, Jaime Prilusky, Hannah Campbell, Alexander Berchansky, Angel Herraez