We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
From Proteopedia
proteopedia linkproteopedia link
|
|
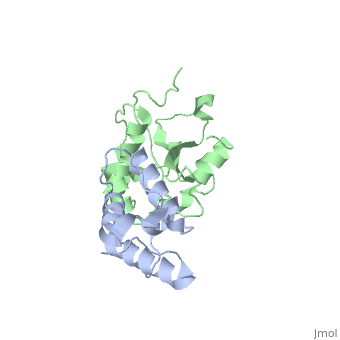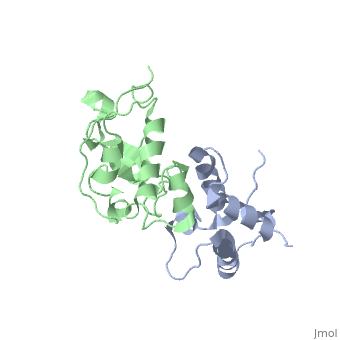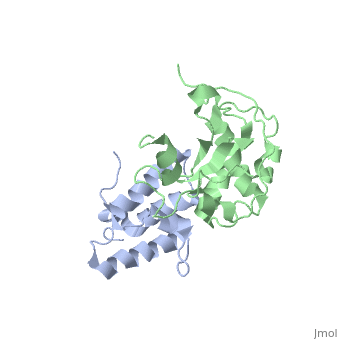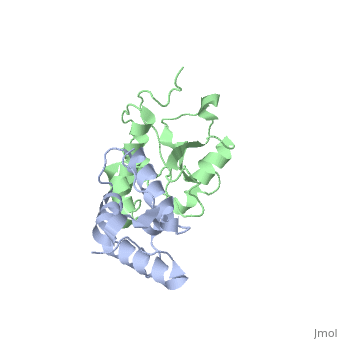
| 2k5x, 1 NMR models ()
|
| Gene:
| imm, ceiE9 (Escherichia coli), col, cei (Escherichia coli)
|
| Related:
| 1imq, 1fsj, 1emv
|
|
|
|
|
| Resources:
| FirstGlance, OCA, PDBsum, RCSB
|
| Coordinates:
| save as pdb, mmCIF, xml
|
Colicins are a type of bacteriocin - peptide and protein antibiotics released by bacteria to kill other bacteria of the same species. Bacteriocins are named after their species of origin; colicins are so-called because they are produced by E. Coli. Because of their narrow killing spectrum which focuses primarily on the species which has made the peptide, bacteriocins are important in microbial biodiversity and the stable co-existence of the bacterial populations.
Colicin peptides are plasmid-encoded. The peptide is released by the cell into the area surrounding it, and then parasitises proteins present in the host cell membrane to translocate across into the host cell. Many protein-protein interactions are involved in the cell entry, and the main system is involved in the grouping of colicins into two families: Group A colicins use the Tol system to enter the host cell, and Group B use the Ton system. Once inside the host cell, the cell killing follows 1st order kinetics - ie one molecule is theoretically sufficient to kill the cell.
The structure of all colicins, of which over 20 have been identified, follows a 3 domain design:
At the N terminus is the Translocation domain (T-)
The Receptor binding domain is at the centre of the peptide (R-)
The C terminus contains the Cytotoxic domain (C-).
Synthesis and Production
Release
Targeting and Receptors
Colicin Uptake
Killing Activities
List of colicins, with their translocation proteins and cytotoxic activity
| Colicin | Group | OM Receptor | Translocation Proteins | Cytotoxic activity
|
| Col A | A | BtuB | OmpF/TolQRAB | Pore-forming
|
| Col E1 | A | BtuB | TolC/TolAQ | Pore-forming
|
| Col E2 | A | BtuB | OmpF/TolQRAB | DNase
|
| Col E3 | A | BtuB | OmpF/TolQRAB | 16s rRNase
|
| Col E4 | A | BtuB | OmpF/TolQRAB | 16s rRNase
|
| Col E5 | A | BtuB | OmpF/TolQRAB | tRNase
|
| Col E6 | A | BtuB | OmpF/TolQRAB | 16s rRNase
|
| Col E7 | A | BtuB | OmpF/TolQRAB | DNase
|
| Col E8 | A | BtuB | OmpF/TolQRAB | DNase
|
| Col E9 | A | BtuB | OmpF/TolQRAB | DNase
|
| Col N | A | OmpF | OmpF/TolQRA | Pore-forming
|
| Col S4 | A | OmpW | OmpF/TolQRAB | Pore-forming
|
| Col K | A | Tsx | OmpF/TolQRAB | Pore-forming
|
| Cloacin DF 13 | A | lutA | TolQRA | 16s rRNase
|
| Col U | A | ? | OmpAF, TolQRAB | Pore-forming
|
| Col 5 | B | Tsx | TolC/TonB, ExbBD | Pore-forming
|
| Col 6 | B | Tsx | TolC/TonB, ExbBD | Pore-forming
|
| Col 7 | B | Tsx | TolC/TonB, ExbBD | Pore-forming
|
| Col 8 | B | Tsx | TolC/TonB, ExbBD | Pore-forming
|
| Col 9 | B | Tsx | TolC/TonB, ExbBD | Pore-forming
|
| Col 10 | B | Tsx | TolC/TonB, ExbBD | Pore-forming
|
| Col-la | B | Cir | Cir/TonB, ExbBD | Pore-forming
|
| Col-lb | B | Cir | Cir/TonB, ExbBD | Pore-forming
|
| Col B | B | FepA | ?/TonB, ExbBD | Pore-forming
|
| Col D | B | FepA | ?/TonB, ExbBD | tRNase
|
| Col M | B | FhuA | ?/TonB, ExbBD | Inhibition of PG synthesis
|
| Col V | B | Cir? | TonB, ExbB | Disruption of membrane potential
|
| Col Js | B | CjrBC | ExbBD, VirB | ?
|
| Col Y | ? | ? | ? | Pore-forming
|