Colicin
From Proteopedia
| |||||||||
| 2k5x, 1 NMR models () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene: | imm, ceiE9 (Escherichia coli), col, cei (Escherichia coli) | ||||||||
| Related: | 1imq, 1fsj, 1emv | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Colicins are a type of bacteriocin - peptide and protein antibiotics released by bacteria to kill other bacteria of the same species. Bacteriocins are named after their species of origin; colicins are so-called because they are produced by E. Coli[1]. Because of their narrow killing spectrum which focuses primarily on the species which has made the peptide, bacteriocins are important in microbial biodiversity and the stable co-existence of the bacterial populations.
Colicin peptides are plasmid-encoded. The peptide is released by the cell into the area surrounding it, and then parasitises proteins present in the host cell membrane to translocate across into the host cell. Many protein-protein interactions are involved in the cell entry, and the main system is involved in the grouping of colicins into two families: Group A colicins use the Tol system to enter the host cell, and Group B use the Ton system. Once inside the host cell, the cell killing follows 1st order kinetics - ie one molecule is theoretically sufficient to kill the cell.
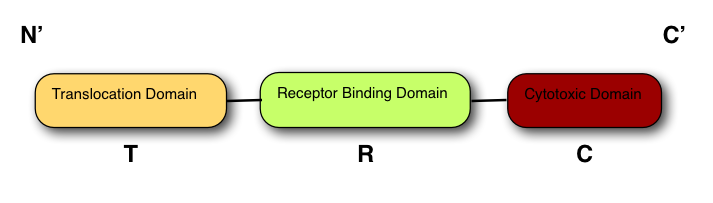
The structure of all colicins, of which over 20 have been identified, follows a 3 domain design:
At the N terminus is the Translocation domain (T-)
The Receptor binding domain is at the centre of the peptide (R-)
The C terminus contains the Cytotoxic domain (C-).
Contents |
Synthesis and Production
Release
Targeting and Receptors
Colicins vary significantly in the receptors that they target to initiate their uptake. The majority of the group A colicins use the BtuB receptor, which is present on E. coli as a vitamin B12 uptake receptor.
Understanding how the colicins can cross the membrane is highly important, as if this could be targeted and exploited it could be useful for novel therapeutic agents. [2]
Colicin Uptake
Colicins are divided into two groups depending on the method of uptake which they target. Group A colicins use the Tol system to bind to and enter the target cell, and group B use the Ton system.
Killing Activities
Colicins kill their target cell through a variety of different methods. The main killing activities are carried out through Pore Formation, and 16s rRNase activity.
The killing activities carried out by colicins could be used medicinally as an alternative to antibiotics in the case where the specific strain of E. coli can be identified. [3]
| Colicin | Group | OM Receptor | Translocation Proteins | Cytotoxic activity |
|---|---|---|---|---|
| Colicin A | A | BtuB | OmpF/TolQRAB | Pore-forming |
| Colicin E1 | A | BtuB | TolC/TolAQ | Pore-forming |
| Colicin E2 | A | BtuB | OmpF/TolQRAB | DNase |
| Colicin E3 | A | BtuB | OmpF/TolQRAB | 16s rRNase |
| Colicin E4 | A | BtuB | OmpF/TolQRAB | 16s rRNase |
| Colicin E5 | A | BtuB | OmpF/TolQRAB | tRNase |
| Colicin E6 | A | BtuB | OmpF/TolQRAB | 16s rRNase |
| Colicin E7 | A | BtuB | OmpF/TolQRAB | DNase |
| Colicin E8 | A | BtuB | OmpF/TolQRAB | DNase |
| Colicin E9 | A | BtuB | OmpF/TolQRAB | DNase |
| Colicin N | A | OmpF | OmpF/TolQRA | Pore-forming |
| Colicin S4 | A | OmpW | OmpF/TolQRAB | Pore-forming |
| Colicin K | A | Tsx | OmpF/TolQRAB | Pore-forming |
| Cloacin DF13 | A | lutA [4] | TolQRA [5] | 16s rRNase [6] |
| Colicin U | A | ? | OmpAF, TolQRAB | Pore-forming |
| Colicin 5 | B | Tsx | TolC/TonB, ExbBD | Pore-forming |
| Colicin 6 | B | Tsx | TolC/TonB, ExbBD | Pore-forming |
| Colicin 7 | B | Tsx | TolC/TonB, ExbBD | Pore-forming |
| Colicin 8 | B | Tsx | TolC/TonB, ExbBD | Pore-forming |
| Colicin 9 | B | Tsx | TolC/TonB, ExbBD | Pore-forming |
| Colicin 10 | B | Tsx | TolC/TonB, ExbBD | Pore-forming |
| Colicin Ia | B | Cir | Cir/TonB, ExbBD | Pore-forming |
| Colicin Ib | B | Cir | Cir/TonB, ExbBD | Pore-forming |
| Colicin B | B | FepA | ?/TonB, ExbBD | Pore-forming |
| Colicin D | B | FepA | ?/TonB, ExbBD | tRNase |
| Colicin M | B | FhuA | ?/TonB, ExbBD | Inhibition of PG synthesis |
| Colicin V | B | Cir? [7] | TonB, ExbB | Disruption of membrane potential |
| Colicin Js | B | CjrBC [8] | ExbBD, VirB [9] | ? |
| Colicin Y | ? | ? | ? | Pore-forming [10] |
Table taken from [11] except where indicated.
References
- ↑ Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, Riley M, Slatin S, Cavard D. Colicin biology. Microbiol Mol Biol Rev. 2007 Mar;71(1):158-229. PMID:17347522 doi:10.1128/MMBR.00036-06
- ↑ Mosbahi K, Walker D, Lea E, Moore GR, James R, Kleanthous C. Destabilization of the colicin E9 Endonuclease domain by interaction with negatively charged phospholipids: implications for colicin translocation into bacteria. J Biol Chem. 2004 May 21;279(21):22145-51. Epub 2004 Mar 23. PMID:15044477 doi:10.1074/jbc.M400402200
- ↑ Cutler SA, Lonergan SM, Cornick N, Johnson AK, Stahl CH. Dietary inclusion of colicin e1 is effective in preventing postweaning diarrhea caused by F18-positive Escherichia coli in pigs. Antimicrob Agents Chemother. 2007 Nov;51(11):3830-5. Epub 2007 Aug 27. PMID:17724148 doi:10.1128/AAC.00360-07
- ↑ Wooldridge KG, Williams PH. Sensitivity of Escherichia coli to cloacin DF13 involves the major outer membrane protein OmpF. J Bacteriol. 1991 Apr;173(8):2420-4. PMID:2013565
- ↑ Thomas JA, Valvano MA. Role of tol genes in cloacin DF13 susceptibility of Escherichia coli K-12 strains expressing the cloacin DF13-aerobactin receptor IutA. J Bacteriol. 1993 Jan;175(2):548-52. PMID:8419302
- ↑ Baan RA, Duijfjes JJ, van Leerdam E, van Knippenberg PH, Bosch L. Specific in situ cleavage of 16S ribosomal RNA of Escherichia coli interferes with the function of initiation factor IF-1. Proc Natl Acad Sci U S A. 1976 Mar;73(3):702-6. PMID:768982
- ↑ Waters VL, Crosa JH. Colicin V virulence plasmids. Microbiol Rev. 1991 Sep;55(3):437-50. PMID:1943995
- ↑ Braun V, Patzer SI, Hantke K. Ton-dependent colicins and microcins: modular design and evolution. Biochimie. 2002 May-Jun;84(5-6):365-80. PMID:12423780
- ↑ Smajs D, Weinstock GM. The iron- and temperature-regulated cjrBC genes of Shigella and enteroinvasive Escherichia coli strains code for colicin Js uptake. J Bacteriol. 2001 Jul;183(13):3958-66. PMID:11395459 doi:10.1128/JB.183.13.3958-3966.2001
- ↑ Riley MA, Cadavid L, Collett MS, Neely MN, Adams MD, Phillips CM, Neel JV, Friedman D. The newly characterized colicin Y provides evidence of positive selection in pore-former colicin diversification. Microbiology. 2000 Jul;146 ( Pt 7):1671-7. PMID:10878131
- ↑ Kleanthous C. Swimming against the tide: progress and challenges in our understanding of colicin translocation. Nat Rev Microbiol. 2010 Dec;8(12):843-8. Epub 2010 Nov 9. PMID:21060316 doi:10.1038/nrmicro2454
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Gemma McGoldrick, Alexander Berchansky, Jaime Prilusky