Ribonuclease A Catalysis
From Proteopedia
| This Sandbox is Reserved from Feb 02, 2011, through Jul 31, 2011 for use by the Biochemistry II class at the Butler University at Indianapolis, IN USA taught by R. Jeremy Johnson. This reservation includes Sandbox Reserved 191 through Sandbox Reserved 200. |
To get started:
More help: Help:Editing |
Contents |
Ribonuclease A Catalysis
Introduction
| |||||||||
| 7rsa, resolution 1.26Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||||
| Activity: | Pancreatic ribonuclease, with EC number 3.1.27.5 | ||||||||
| |||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Acid Base Catalysis
In organic chemistry acid/base catalysis is the addition of an acid or base to accelerate a chemical reaction. Ribonuclease A, (RNase A), also uses acid/base catatalysis to chemically change its substrates. Acidic or basic residues of the enzyme transfer protons to or from the reactant in order to stabilize the developing charges in the transition state. The transfer of protons usually creates better leaving groups, making the reaction more energetically favorable. Histidine is a very common amino acid residue involved in cataylsis, as histidine has a pKa value close to neutral, (pKa=6); therefore, histidine can both accept and donate protons at physiological pH.
Acid/base catalysis by an enzyme is dependent on the pH of the environment and the pKa's of their residues. The pKa value will increase for an acidic residue if the environment is hydrophobic or if the adjacent residues are of similar charges. In the same environmental conditions, a basic residue will decrease the pKa. Having the ability to alter the pKa of certain residues such as Histidine, increases the diversity of reactions that can take place (reword).
Active Site Structure
RNase A uses acid/base catlysis to speed up RNA hydrolysis. This occurs in the which is found in the cleft of RNase A and is the location of the chemical change in bound substrates. Subsites lining the active site cleft are important to the binding of single stranded RNA. Large quantities of positively charged residues, such as and and , recognize the negative charge on the phosphate back bone of the RNA strand.
The active site for RNase A, although fairly nonspecific, has some specificity for sites RNA hydrolysis. , located next to the active site, will hydrogen bond to pyrimidine bases, but sterically hinder the binding of a purine on the 5' strand of OH. Thr45 significantly decreases the rate of hydrolysis of polymeric purine strands, such as poly A, by a thousand fold, as compared to polymeric pyrimidine strands.
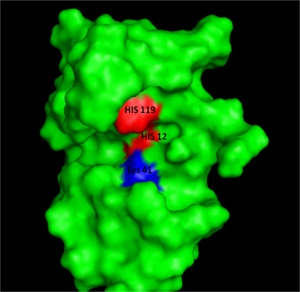
Early studies on RNase A catalysis showed that alkylation of His12 and His119 significantly decreased its catalytic activity, prompting the hypothesis that these two histidines were the acid/base catalyst. Confirmation of this hypothesis came when these histidines were replaced with alanine and the reaction rates of either mutation dropped by ten-thousand fold.
Acid Base Catalysis by RNase A
Mechanism
RNase A catalyzes the cleavage of the Phosphodiester bonds in two steps: the formation of the pentavalent phosphate transition state and subsequent degradation of the 2’3’ cyclic phosphate intermediate. An important part of the reaction is the ability of histidine (His 12 and His119) to both accept and donate electrons, allowing these histidine to be an acid or a base, making the reaction pH dependent [1]Raines, R. Ribonuclease A. Chemistry Review: (1998) Vol. 98 pp. 1045-1068
2. </ref>Turcotte, R., Raines, R., Interactions of Onconase with Human Ribonuclease Inhibitor. Biochemical Biophysical Research Communities:(2008) Vol. 377, Iss. 4, pp. 512-414
3. </ref>Vicentini,A., Protein Chemical and Kinetic Characterization of Recombinant Porcine Ribonuclease Inhibitor Expressed in Saccharomyces cerevisae. Biochemistry: (1996) Vol. 29, pp.8827-8834
4. </ref>Wlodrawer, A., Svensson, L., Sjohin, L., Gilliland, G. Structure of Phosphate-Free Ribonuclease A Refined at 1.26A. Biochemistry:(1988) Vol. 27 pp. 2705-2717
Related Sites
http://en.wikipedia.org/wiki/RNase_A
http://en.wikipedia.org/wiki/Ribonuclease_inhibitor
Proteopedia Page Contributors and Editors (what is this?)
Nathan Clarke, David Canner, Alexander Berchansky, R. Jeremy Johnson, OCA