Alcohol dehydrogenase
From Proteopedia
| |||||||||
| 1htb, resolution 2.40Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , , | ||||||||
| Gene: | HUMAN BETA3 CDNA (Homo sapiens) | ||||||||
| Activity: | Alcohol dehydrogenase, with EC number 1.1.1.1 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Alcohol dehydrogenase (PDB id 1htb), ADH, is an 80kDa enzyme that catalyzes the 4th step in the metabolism of fructose before glycolysis. In the 4th step, glyceraldehyde is converted to the glycolytic intermediate DHAP by the NADH-dependent, ADH catalyzed reduction to glycerol.[1] ADH catalyzes the oxidation of primary and secondary alcohols to their corresponding aldehydes and ketones through a mechanism that involves the removal of a hydrogen.
Contents |
Structure
The initial scene () shows an overview of the molecule, allowing for a general look at the tertiary structure of alcohol dehydrogenase (it is complexed with Cl, Pyz, NAD, and Zn). A second scene () shows a close view of the ligand within each subunit. Labels have been placed on NAD, CL, and Zn to clearly establish the structure.
Within alcohol dehydrogenase, site of alcohol dehydrogenase has three important residues, Phe 93, Leu 57, and Leu 116. These three residues work together to bind to the alcohol substrate.[2]
Zn plays an important role in the catalysis. It funtions by electrostatically stabilizing the oxygen in alcohol during the reaction, which causes the alcohol to be more acidic. At the , Zinc coordinates with Cys 146, Cys 174, and His 67.[3]
NAD functions as a cosubstrate in the dehydration. NAD binds to numerous residues in a series of beta-alpha-beta folds. shows the domain where NAD binds, and many of the residues with which it interacts are selected.
[4]
Alcohol dehydrogenase exists as a dimer with a zinc molecule complexed in each of the subunits. It has a SCOP catagory of an alpha and beta protein. At the N-terminal, there is a domain that is all beta; however, the C-Terminal domain is alpha and beta, so the catagory is alpha and beta. The C-Terminal core has 3 layers of alpha/beta/alpha and parallel beta sheets of 6 strands.[5]
Reaction and Mechanism
In the oxidation mechanism, ADH is momentarily associated with nicontinamide adenine dinucleotide (NAD+), which functions as a cosubstrate. In its reaction, alcohol dehydrogenase uses zinc and NAD to facilitate the reaction. The function of zinc is to position the –OH group on the ethanol in a conformation that allows for the oxidation to occur. NAD then acts as a cosubstrate and performs the oxidation.
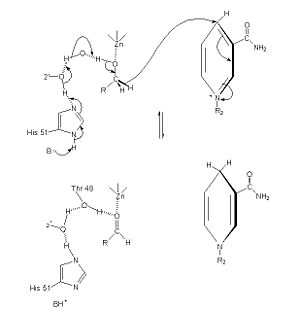
[6] The of alcohol dehydrogenase reaction is as follows: CH3CH2OH + NAD+ -> CH3COH (acetaldehyde) + NADH + H+ (Note: The reaction is actually reversible although the arrow does not show it) [7] The step-wise reduction mechanism for ADH is shown on the left. In the mechanism, His 51 is deprotonated and activated by a base catalyst. This allows histidine to accept a proton from NAD, which also draws a proton Thr 48. As a result of the proton transfer, the Thr is prepared to accept a proton from the alcohol substrate. While Thr accepts the proton, there is also a hydride transfer to NAD. The whole process can be summarized as the oxidation of an alcohol to an aldehyde in concert with the transfer of a hydride to NAD.[8]
The Mechanism for alcohol dehydrogenase follows an random bisubstrate mechanism.[9] In the mechanism, the NAD+ and alcohol bind to the enzyme, so that the enzyme is now attached to the two subtrates. While attached, the hydrogen is formally transferred from the alcohol to NAD, resulting in the products NADH and a ketone or aldehyde. The two products are then released, and the enzyme has catalyzed the reaction.
Kinetics
The alcohol dehydrogenase catalyzed aldehyde-NADH reaction show kinetics consistent with a random-order mechanism, and the rate-limiting step is the dissociation of the product enzyme-NAD+ complex. [10] Alcohol dehydrogenase is more effective for smaller alcohol substrates, and it becomes less effective as substrate size increases. It is also more effective for primary than secondary alcohols.[11] In a study where ADH was immobilized in tresyl-chloride-activate agarose, it was shown that the Michaelis-Menten model could not take into consideration all the constraints induced by the immobilization on the enzyme properties but that the Theorell-Chance model was more appropriate.[12]
Regulation
Substrate size is a regulator, where larger substrates inhibit alcohol dehydrogenase. Further, alcohol dehydrogenase is somewhat inhibited if the substrate is a secondary alcohol, as opposed to a primary alcohol. [13] Pyrazoles have also been shown to be inhibitors of ADH. [14] Other inhibitors include heavy metals, thiourea, purine and pyrimidine derivatives, and both chloroethanol and flouroethanol. [15] Activators include sulfhydryl activating reagents, mercaptoethanol, dithiothreitol, and cysteine.[16]
Additional Resources
For additional information, see: Carbohydrate Metabolism
3D Structures of Alcohol dehydrogenase
ADH I
3jv7 – RrADH I – Rhodococcus rubber
2vna - hADH I catalytic domain - human
2hcy – yADH I – yeast
ADH I binary complex
1u3t – hADH I α chain + inhibitor
1hsz, 1hdz, 1hud - hADH I β chain + NAD
1u3w - hADH I γ chain + inhibitor
1ht0 - hADH I γ chain (mutant) + NAD
ADH I ternary complex
2xaa – RrADH I + NAD + alcohol
3fx4 – ADH I + NADP + inhibitor – pig
2w98, 2w4q – hADH I catalytic domain + NADP + inhibitor
1hso - hADH I α chain + NAD + pyrazole derivative
1hdx - hADH I β chain + NAD + alcohol
1u3u, 1u3v - hADH I β chain + inhibitor
1deh, 1hdy - hADH I β chain + NAD + pyrazole derivative
1htb - hADH I β3 chain + NAD + pyrazole derivative
ADH II
3owo – ZmADH II iron-dependent – Zymomonas mobilis
ADH II binary complex
3ox4 - ZmADH II iron-dependent + NAD
3cos - hADH II + NAD + Zn
1e3e – mADH II + NADH – mouse
1e3l - mADH II (mutant) + NADH
1e3i - mADH II + NADH + inhibitor
ADH IV
1ye3, 8adh, 5adh - hoADH IV e chain – horse
1qlj - hoADH IV e chain (mutant)
3iv7 – ADH IV – Corynebacterium glutamicum
ADH IV binary complex
2jhf, 2jhg, 1het, 1heu, 1hf3, 1ee2, 1oxi, 2ohx, 6adh - hoADH IV e chain + NAD
1adb, 1adc, 1adf, 1adg, 7adh - hoADH IV e chain + NAD derivative
1mgo, 1ju9, 1qlh, 1a72 - hoADH IV e chain (mutant) + NAD
1d1s, [[1agn – hADH IV σ chain + NAD
1d1t - hADH IV σ chain (mutant) + NAD
ADH IV ternary complex
3oq6, 1qv6, 1qv7, 1a71, 1axe, 1axg – hoADH IV e chain (mutant) + NAD + alcohol
1p1r, 1ldy, 1lde - hoADH IV e chain + NADH + formamide derivative
1n92 - hoADH IV e chain + NAD + pyrazole derivative
1bto, 3bto - hoADH IV e chain + NADH + butylthiolane derivative
1n8k - hoADH IV e chain (mutant) + NAD + pyrazole
1mg0, 1hld - hoADH IV e chain + NAD + alcohol
ADH
1a4u – SlADH – Scaptodrosophila lebanonensis
3my7 – ADH ACDH domain – Vibrio parahaemolyticus
3meq – ADH – Brucella suis
3l4p – ADH – Desulfovibrio gigas
1jvb - SsADH – Sulfolobus solfataricus
3i4c, 1nto, 1nvg – SsADH (mutant)
3goh – ADH – Shewanella oneidensis
3gaz – ADH residues 2-334 – Novosphingobium aromaticivorans
2eih – ADH – Thermus thermophilus
1rjw – ADH – Geobacillus stearothermophilus
1vj0, 1vhd – TmADH
ADH binary complex
3l77 – ADH short-chain + NADP – Thermococcus sibiricus
1h2b – ADH + NAD – Aeropyrum pernix
1f8f – Benzyl-ADH + NAD – Acinetobacter calcoaceticus
1o2d - TmADH + NADP – Thermotoga maritime
1b16, 1b14, 1b15 - SlADH + NAD derivative
1cdo – ADH + NAD - cod
1rhc – ADH F420-dependent +F420-acetone – Methanoculleus thermophilus
ADH ternary complex
1mg5 – ADH + NADH + acetate – Drosophila melanogaster
1r37 – SsADH + NAD + alcohol
1sby – SlADH + NAD + alcohol
1b2l - SlADH + NAD + cyclohexanone
1llu - ADH + NAD + alcohol – Pseudomonas aeruginosa
NADP-dependent ADH
1ped - CbADH – Clostridium beijerinckii
2b83, 1jqb – CbADH (mutant)
2nvb - TbADH (mutant) – Thermoanaerobacter brockii
3ftn, 3fpc, 3fpl, 3fsr – ADH chimera
1y9a - EhADH – Entamoeba histolytica
[[2oui – EhADH (mutant)
1kev – CbADH + NADPH
1bxz – CbADH catalytic domain + alcohol
1ykf – TbADH + NADP
1p0c – RpADH8 – Rana perezi
1p0f – RpADH + NADP
R-specific ADH
1nxq - LbRADH – Lactobacillus brevis
1zk2, 1zk3 - LbRADH (mutant)
1zjy, 1zjz, 1zk0, 1zk1 – LbRADH (mutant) + NADH + alcohol
1zk4 - LbRADH (mutant) + NADH + acetophenone
Specific alcohol ADH
2cf5, 2cf6 – Cinnamyl-ADH – Arabidopsis thaliana
1piw, 1q1n, 1ps0 – Cinnamyl-yADH
1m2w – Mannitol-ADH – Pseudomonas fluorescens
1w6s – Methanol-ADH – Methylobacterium extorquens
1yqx – Sinapyl-aADH II – aspen
1yqd – Sinapyl-aADH II + NADP
1bdb – Biphenyl dihydrodiol-ADH + NAD - Pseudomonas
Quinohemoprotein ADH
1kv9, 1yiq – PpQADH II + PQQ + heme – Pseudomonas putida
1kb0 - QADH I + PQQ + heme – Comamonas testosteroni
Hydroxyacyl-CoA dehydrogenase
2et6 – HADH – Candida tropicalis
1zcj – rHADH - rat
1e3s – rHADH II + NADH
1gz6 - rHADH II residues 1-319 + NADH
1e3w - rHADH II + NADH + keto butyrate
1e6w - rHADH II + NADH + alcohol
1lsj, 1lso– hHADH (mutant) + NAD
References
- ↑ Voet, et. al. Fundamentals of Biochemistry: 3rd Edition. Hoboken: Wiley & Sons, Inc, 2008.
- ↑ Protein: Alcohol Dehydrogenase. The College of Saint Benedict and Saint John's University. 1 March 2010 < http://www.users.csbsju.edu/~hjakubow/classes/rasmolchime/99ch331proj/alcoholdehydro/index.htm>
- ↑ Protein: Alcohol Dehydrogenase. The College of Saint Benedict and Saint John's University. 1 March 2010 < http://www.users.csbsju.edu/~hjakubow/classes/rasmolchime/99ch331proj/alcoholdehydro/index.htm>
- ↑ Protein: Alcohol Dehydrogenase. The College of Saint Benedict and Saint John's University. 1 March 2010 < http://www.users.csbsju.edu/~hjakubow/classes/rasmolchime/99ch331proj/alcoholdehydro/index.htm>
- ↑ Protein: Alcohol dehydrogenase from Human (Homo sapiens), different isozymes. SCOP. 2009. 1 March 2010 < http://scop.berkeley.edu/data/scop.b.d.c.b.b.c.html>
- ↑ Protein: Alcohol Dehydrogenase. The College of Saint Benedict and Saint John's University. 1 March 2010 < http://www.users.csbsju.edu/~hjakubow/classes/rasmolchime/99ch331proj/alcoholdehydro/index.htm>
- ↑ Voet, et. al. Fundamentals of Biochemistry: 3rd Edition. Hoboken: Wiley & Sons, Inc, 2008.
- ↑ Protein: Alcohol Dehydrogenase. The College of Saint Benedict and Saint John's University. 1 March 2010 < http://www.users.csbsju.edu/~hjakubow/classes/rasmolchime/99ch331proj/alcoholdehydro/index.htm>
- ↑ Voet, et. al. Fundamentals of Biochemistry: 3rd Edition. Hoboken: Wiley & Sons, Inc, 2008.
- ↑ Dickinson FM, Monger GP. A study of the kinetics and mechanism of yeast alcohol dehydrogenase with a variety of substrates. Biochem J. 1973 Feb;131(2):261-70. PMID:4352908
- ↑ Dickinson FM, Monger GP. A study of the kinetics and mechanism of yeast alcohol dehydrogenase with a variety of substrates. Biochem J. 1973 Feb;131(2):261-70. PMID:4352908
- ↑ Bille V, Remacle J. Simple-kinetic descriptions of alcohol dehydrogenase after immobilization on tresyl-chloride-activated agarose. Eur J Biochem. 1986 Oct 15;160(2):343-8. PMID:3769934
- ↑ Dickinson FM, Monger GP. A study of the kinetics and mechanism of yeast alcohol dehydrogenase with a variety of substrates. Biochem J. 1973 Feb;131(2):261-70. PMID:4352908
- ↑ Blomstrand R, Ostling-Wintzell H, Lof A, McMartin K, Tolf BR, Hedstrom KG. Pyrazoles as inhibitors of alcohol oxidation and as important tools in alcohol research: an approach to therapy against methanol poisoning. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3499-503. PMID:115004
- ↑ Alcohol Dehydrogenase. Worthington Biochemical Corporation . 31 March 2010 < http://http://www.worthington-biochem.com/ADH/default.html>
- ↑ Alcohol Dehydrogenase.Worthington Biochemical Corporation . 31 March 2010 < http://http://www.worthington-biochem.com/ADH/default.html>
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, David Canner, Joel L. Sussman, David Birrer