User:Jamie Abbott/Sandbox1
From Proteopedia
Contents |
Histone Acetyltransferase GCN5
Histone Acetyltransferase (HAT) GCN5 is a ~94 kD (837 amino acid) protein. It is a nuclear HAT or A-type HAT. GCN5 belongs to the GCN5-related N-acetyltransferase (GNAT) superfamily that includes the HATs, aminoglycoside N-acetyltransferases, mycothiol synthase, protein N-myristoyltransferase, and the Fem family of amino acyl transferases.[1] Most if not all HATs function in vivo as members of often large multisubunit complexes, many of which were initially characterized as transcriptional regulators. GCN5 has been shown to be part of the STAGA (SPT3-TAFII31-GCN5-L acetylase)[2] complex as well as the TFTC (TATA-binding protein-free TAFII containing)[3] complex.
GCN5 catalyzes the acetylation of specific Lysine residues of histones H3 and H4. More specifically GCN5 is know to acetylate the lysine residues at position 8 and 16 of H4 and 14 of H3 in vivtro. [4]. Acetylation results in the neutralization of charged lysine residues which is hypothesized to weaken histone:DNA contacts[5] as well as alter histone:histone interactions[6]. Chromatin modification more specifically reversible histone acetylation has been associated with gene activation and consequently transcriptional activity for many years.
HAT Domain
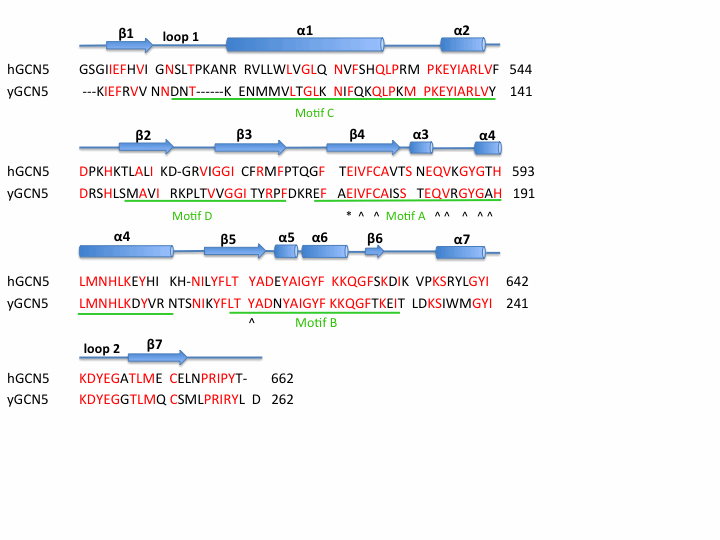
Template:STRUCTURE 1z4r The HAT domain of human GCN5 [7] consists of amino acid residues 496-658 with mixed α/β topology. This mixed α/β structure consists of 7 and 7 anti-parallel . AcCoA is bound via hydrogen bonds in a cleft on the surface of the protein. Residues involved in AcCoA include Val587, Gly589, Gly591, Thr592, Cys579, and Tyr617. There are two long flexible loops in the HAT domain. links helix α1 and strand β1. connects helix α7 and strand β7 and has been associated with substrate binding and specificity. [8] [9]
|
Catalysis
GCN5 catalyzes the transfer of an acetyl group from acetyl coenzyme A () onto the ε-amino group of specific lysine residues present in the amino-terminal tails of each of the core histones, H3 and H4, resulting in the neutralization of a single positive charge. [10] Currently, it has been demonstrated that the catalytic mechanism for yeast GCN5 involves a glutamic acid -173 residue, that acts as a general base. The Glu173 residue must deprotonate the ε-amino group of Lys14 of histone H3 prior to attack on the carbonyl carbon of AcCoA. [11] Comparing the sequence of human GCN5 HAT domain with yeast GCN5 strongly suggests that the catalytic mechanism of acetylation would be very similar. The conserved glutamic acid -173 of yeast GCN5 aligns with glutamic acid -575 of human GCN5 and therefore most likely functions as the general base in acetylation of histones H3 and H4.
Bromo Domain
Template:STRUCTURE 3d7c The bromodomain is a highly conserved domain found to be a part of many chromatin remodeling proteins and nearly all HAT structures contain a bromodomain. This 110 amino acid motif was originally identified as a sequence motif common to the Drosophila brahma and female-sterile homeotic proteins, the yeast SWI2/SNF2 proteins and the human CCG1 protein [12][13] While the bromo domain is fairly close to the HAT domain of GCN5 no evidence suggests that it is involved in or necessary for histone acetyl transferase activity.
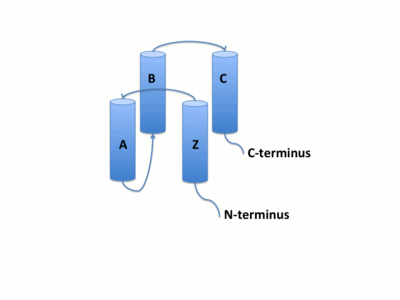
The GCN5 bromor domain is 71% α-helical with no β structure. The bromodoamin is 110 amino acids and forms a (αZ,αA,αB,and αC). Helices αZ and αA are connected by the ZA loop while helices αB and αC are connected by the BC loop. The up-and-down four-helix bundle of the GCN5 bromodomain has left handed topology as a result of the orientation of the long ZA loop. Loops ZA and BC pack together to form a hydrophobic pocket that may be involved in protein-protein interactions.[14]
The bromodomain is separated from the HAT domain by 57 amino acids residues which contain an ADA2 interaction domain.[15]
| [[Image:Electrostatic map I.jpg|center|thumb|upright|Hydrophobic Pocket formed by AZ and BC Loops]
|
|
Protein-Protein Interactions
Salt Bridge Formation
PCAF_N Domain
Structural Prediction
Post-Translational Modifications
Evoultionary Conservation
References
- ↑ Vetting MW, S de Carvalho LP, Yu M, Hegde SS, Magnet S, Roderick SL, Blanchard JS. Structure and functions of the GNAT superfamily of acetyltransferases. Arch Biochem Biophys. 2005 Jan 1;433(1):212-26. PMID:15581578 doi:10.1016/j.abb.2004.09.003
- ↑ Martinez E, Kundu TK, Fu J, Roeder RG. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. J Biol Chem. 1998 Sep 11;273(37):23781-5. PMID:9726987
- ↑ Brand M, Yamamoto K, Staub A, Tora L. Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J Biol Chem. 1999 Jun 25;274(26):18285-9. PMID:10373431
- ↑ PMID: __________
- ↑ . PMID:5339769
- ↑ Wolffe AP, Hayes JJ. Chromatin disruption and modification. Nucleic Acids Res. 1999 Feb 1;27(3):711-20. PMID:9889264
- ↑ Schuetz A, Bernstein G, Dong A, Antoshenko T, Wu H, Loppnau P, Bochkarev A, Plotnikov AN. Crystal structure of a binary complex between human GCN5 histone acetyltransferase domain and acetyl coenzyme A. Proteins. 2007 Jul 1;68(1):403-7. PMID:17410582 doi:10.1002/prot.21407
- ↑ Clements A, Poux AN, Lo WS, Pillus L, Berger SL, Marmorstein R. Structural basis for histone and phosphohistone binding by the GCN5 histone acetyltransferase. Mol Cell. 2003 Aug;12(2):461-73. PMID:14536085
- ↑ Poux AN, Marmorstein R. Molecular basis for Gcn5/PCAF histone acetyltransferase selectivity for histone and nonhistone substrates. Biochemistry. 2003 Dec 16;42(49):14366-74. PMID:14661947 doi:10.1021/bi035632n
- ↑ Schuetz A, Bernstein G, Dong A, Antoshenko T, Wu H, Loppnau P, Bochkarev A, Plotnikov AN. Crystal structure of a binary complex between human GCN5 histone acetyltransferase domain and acetyl coenzyme A. Proteins. 2007 Jul 1;68(1):403-7. PMID:17410582 doi:10.1002/prot.21407
- ↑ Tanner KG, Langer MR, Kim Y, Denu JM. Kinetic mechanism of the histone acetyltransferase GCN5 from yeast. J Biol Chem. 2000 Jul 21;275(29):22048-55. PMID:10811654 doi:10.1074/jbc.M002893200
- ↑ Owen DJ, Ornaghi P, Yang JC, Lowe N, Evans PR, Ballario P, Neuhaus D, Filetici P, Travers AA. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J. 2000 Nov 15;19(22):6141-9. PMID:11080160 doi:10.1093/emboj/19.22.6141
- ↑ Tamkun JW, Deuring R, Scott MP, Kissinger M, Pattatucci AM, Kaufman TC, Kennison JA. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992 Feb 7;68(3):561-72. PMID:1346755
- ↑ Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999 Jun 3;399(6735):491-6. PMID:10365964 doi:10.1038/20974
- ↑ Candau R, Zhou JX, Allis CD, Berger SL. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 1997 Feb 3;16(3):555-65. PMID:9034338 doi:10.1093/emboj/16.3.555