Ketosteroid Isomerase
From Proteopedia
Contents |
Ketosteroid Isomerase
Introduction
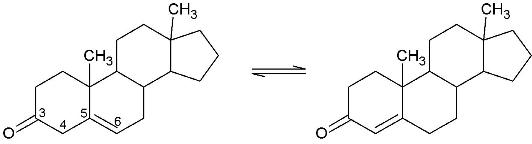
Template:STRUCTURE 1isk (KSI, EC#5.3.3.1) is an enzyme that catalyzes the isomerization of 3-oxo-Δ5 ketosteroids to their hormonally active Δ4-conjugated isomers, as illustrated below.[1], [2]
This reaction is essential in the biosynthesis of steroids in mammals where KSI is a membrane-bound complex.[3] In bacteria, however, KSI exists as a soluble protein is involves in catabolism of steroids.[3] It was first isolated in and has been extensively studied in Commamonas tetosteroni (TI), a bacteria that is capable of growth with testosterone as its sole carbon source.[4] Structural and kinetic studies of this and its homolog from Pseudomonas putida with which it shares 34% sequence and near identical structural homology.[1],[3] It is one of the most efficient known enzymes with an essentially diffusion limited rate of catalysis.[2],[5] It is capable of increasing the catalytic rate by eleven orders of magnitude.[6] The high degree of efficiency is believed to be due to a preference for the transition state to move towards products rather than reactants although the exact mechanism of this preference is unclear.[1] Its high catalytic efficiency and unique active site geometry have made it fertile ground for examining the validity of the low barrier hydrogen bond hypothesis[7] and electrostatic preorganization.[8].
Structure
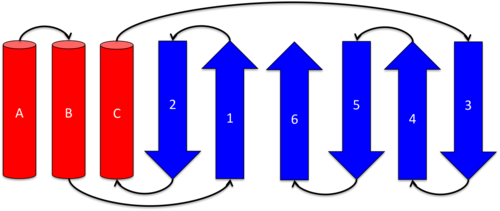
Ketosteroid isomerase exits as a 28 kDa homodimeric protein, in which the two dimers related to each other via hydrophobic and electrostatic interactions.[5] Each monomer consists of a curved and three . These secondary structures define a conical closed barrel geometry, with one open and one closed end, and create a deep pocket in which the active site resides.[3],[9] This unique geometry is shared by several other proteins (scytalone dehydratase, nuclear transport factor 2, and naphthalene 1,2-dioxygenase), however, these molecules do not share functional or sequence homology. It is speculated that this unique protein structure may enable better binding of hydrophobic substrates such as steroids.[3]
Alpha-Helices
Each monomer of KSI contains three α-helices. is contains residues Thr3 to (N-cap) to Ala20. contains residues Asp22(N-cap) to Phe30. contains residues Thr48 to Leu61. [5]
Alpha-Helix Capping Motifs
α-helix capping motifs are defined by specific patterns of hydrophobic interactions and hydrogen bonding that occur at both the initiation and termination of these secondary structural elements. [10] Capping motifs can also help support the formation of tertiary structural elements. KSI contains examples several of these motifs involved in both C-terminal and N-teminal capping, with two illustrations of C-terminal capping motifs as illustrated below (nomenclature adapted from that of Aurora and Rose [10]):
Alpha-L C-terminal capping motif
An α-L C-terminal motif is located at the terminus of helix A (Ala17 to Leu23) and is defined by the following amino acid sequence ALNA-GDLD where Ala20 is in the C-cap position. The α-L motif is stabilized by a hydrogen bond between the backbone carbonyl of Ala17 and the backbone amide hydrogen of Gly21. Furthermore, a hydrophobic interactions between Leu18 and Leu23 adds stability to this structure. The motif is somewhat constrained by the presence of only one residue (Gly21) between helices A and B.
Proline C-terminal capping motif
The presence of a proline at position 62 "breaks" helix C's secondary structure. Leu59 and Val65 form a hydrophobic stable to support this secondary structural element.
Alpha-Helix Packing
Helix B is packed between Helices A and C in a nearly. Helices B and C cross the enzymatically active cavity forming the cavity's "front face."[5]
Beta-Sheet
Each monomer of KSI contains a . Strand 1 of the β-sheet is composed of residues Glu43 to Gly47. Strand 2 is contains residues Ala34 to Asp38 and runs antiparallel to strand 1. Leu63 to Val74 make up strand 3 which runs antiparallel to strand 4 (Glu77 to Tyr88). Residues Arg91 to Phe104 and Lys108 to Gly124 form strands 5 and 6 respectively. Strand 5 runs antiparallel to strand 4, while strand 6 runs antiparallel to strand 5 and parallel to strand 1.[5] Each β-sheet contains two β-bulges: and . Both the β-bulges participate in dimer-dimer interactions, potentially leading to their stabilization. The juxtaposition of the two β-bulges on each side of the sheet with a central proline residue creates a substantial in the β-sheet.[5]
Beta-Turns and Loops
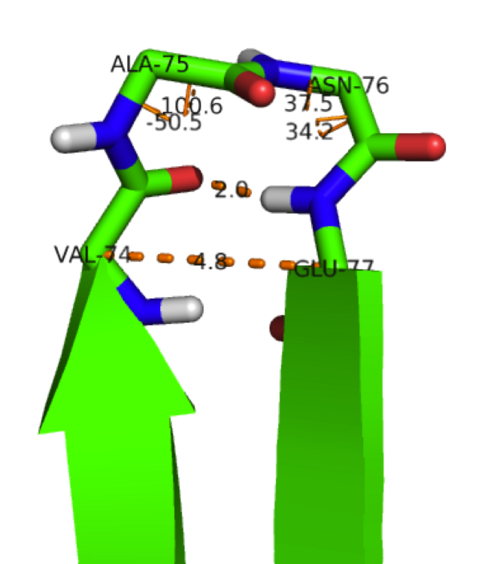
β-strand 1 is connected to β-strand 2 via a four residue linkage (Pro39 to Ser42) with Pro39 being in a cis-conformation. Two examples of type-II β-turns can also be observed in KSI-between β-stands 3 & 4 (Ala75-Asn76→see figure) and β-strands 4 & 5 (Gln89-Gly90). A loop structure (Asn105-Val107) also connects β-strands 5 and 6.
Proteopedia Page Contributors and Editors (what is this?)
Laura M. Haynes, Michal Harel, Joel L. Sussman, Alexander Berchansky