User:Bianca Varney/Replication Terminator Protein
From Proteopedia
|
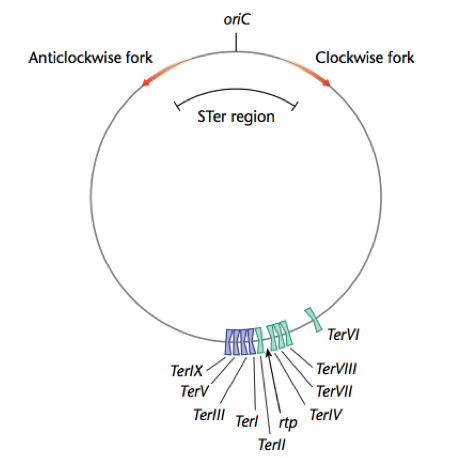
Replication Terminator Protein (RTP) binds bacterial DNA at Termination (Ter) sites that lie opposite the origin of replication (OriC). When the replication forks meet with RTP, which bound to Ter sites, replication is arrested, and DNA polymerase falls off the bacterial chromosome. However, RTP-Ter interactions are orientation specific, and will only arrest the replication forks traveling in on one direction; either clockwise or anticlockwise once each fork has copied more than half the bacterial chromosome. This permits the entire bacterial chromosome to be copied.
Bacterial replication termination has been well studied in E. coli and gram-positive B. subtilis. The proteins involved in this termination process differ structurally in E.coli and B.subtilis, although each contains similar contrahelicase activity and performs similar functions in arresting replication. The replication termini sequences are located in two clusters approximately 1800 from the origin of replication (ori), and orientated such that each cluster has opposite polarity and are therefore inverted repeats. This means that counterclockwise replication forks can move beyond the first cluster of ter sites, the left terminus, but are arrested at the second cluster, the right terminus, containing correct polarity (FIG). Similarly, the clockwise fork will proceed through the first cluster and will be arrested at the second. (CHANGE). The bipartite ter nucleotide sequence is overlapping and each inverted repeat contains a core (IRIB) and an axillary (IRIA) sites. RTP binds to these sequences, resulting in the impediment the replication fork helicase.