User:David McDonald/Replication Termination Protein
From Proteopedia
As with most bacteria, DNA replication of the circular chromosome of B. Subtilis occurs in a bi-directional fashion, starting from a common origin of replication (ori) and ending in the termination region, approximately 180o from the ori. The two replication forks are forced to meet in the termination region by replication termination proteins (RTPs) complexed to specific, unidirectional DNA termination sits (Ter sequences) termination region and arrest the action of the replication fork in a directional manner. That is, there are RTP:Ter complexes which stop replication in the clockwise and in the anti-clockwise direction. The clockwise replication fork is unaffected by the RTP:Ter complexes for the anti-clockwise fork and vice versa.
Contents |
DNA Replication in Bacteria
Structure
|
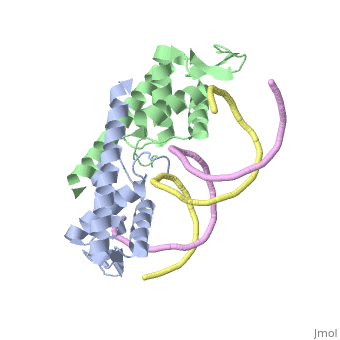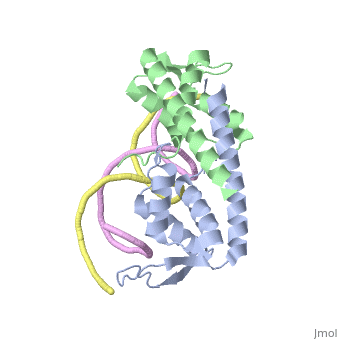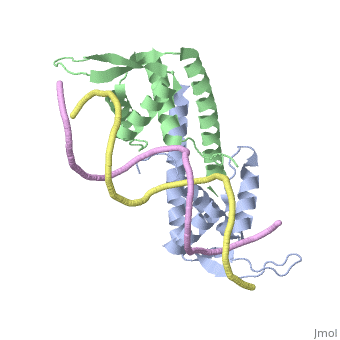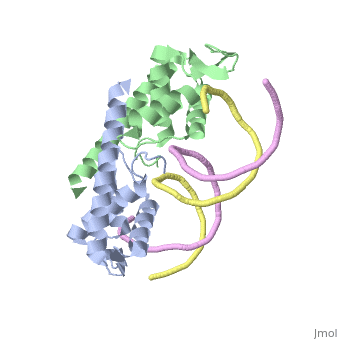
contains 122 amino acid residues and is an example of a winged helix structure, in the α+β protein folding family, containing and (1). In a cell, RTP exists as a homodimer, where the monomer subunits are tightly associated through antiparallel coiled-coil interactions between the Copy and paste the following line where you want the scene link to appear (scroll down if needed) and edit the TextToBeDisplayed:
.
DNA Binding
|
|
Replication Termination
Tus: a homologous Escherichia Coli protein
Works Cited
1. Crystal structure of replication terminator protein of B. subtilis at 2.6 Å. Bussiere, DE, Bastia, D and White, SW. 80, 1995, Cell, pp. 651-660.