Group:MUZIC:Enigma Family
From Proteopedia

Contents |
Enigma family: PDZ- and LIM-domain proteins of the cytoskeleton
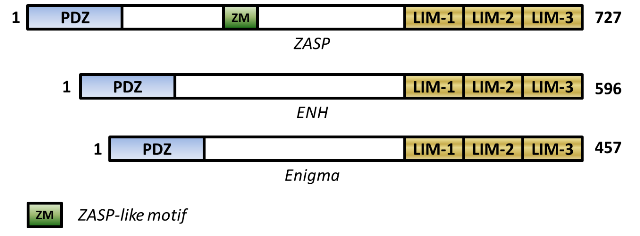
Three member proteins have extensively been described and characterized within this subfamily: Enigma protein, Enigma Homologue (ENH) protein and ZASP/Cypher/Oracle ('ZASP'[1] being the human orthologue of 'Cypher'[2] which is found in mouse and also identified by independent researchers who named it 'Oracle'[3]).
Didactically, protein members of the enigma subfamily typically possess within their structure: (1) an N-terminal PDZ domain (a domain which is named after the first three proteins where it was initially characterized i.e. PSD 95, Disc large protein and Zonula Occludens 1), and (2) three C-terminal LIM domain (a domain which is named after the first three proteins where it was characterized i.e. Lin-11, Isl1 and Mec-3)[4].
The member proteins have all been located to the mammalian muscle cells, some specific to the heart and skeletal muscle Z-disk. They interact via their PDZ domains with motor protein components of the Z-disk and also recruit signalling molecules via their LIM domains or internal motifs, for example ZM motif (ZASP-like motif which is sandwiched between the PDZ- and LIM-domains in ZASP)[5]. These interactions via their PDZ- and LIM-domains have been suggested to be important for targeting/sustaining interacting protein complexes within the sarcomere for a physiologically functional muscle.
Sequence Annotation
Enigma protein alternatively referred to as PDLIM7 (PDZ and LIM domain protein 7) is the first and representative member of the Enigma subfamily. Initially characterized in human as ~49.85 kDa protein with an N-terminal PDZ domain and three C-terminal LIM domains [6], [7]. Five alternatively spliced isoforms are presently identified [1]
Enigma Homologue (ENH) protein is an ~67 kDa, 596-amino-acid human ortholog of rat ENH
protein [8]. ZASP is the 78 kDa, 727-amino-acid human ortholog of Cypher, independently identified in heart and skeletal
muscle[9]. ZASP contains a ZM-motif (ZASp-like motif) which has been identified to interact with the spectrin repeats of a-actinin[10]
The ALP/Enigma subfamily proteins contain one PDZ domain
and one or three LIM domains. . LIM domain(s) of ALP/Enigma subfamily proteins interact
with different domains in various proteins, particularly in signaling
factors. This common structural feature supports the notion
that ALP/Enigma proteins serve as adaptor proteins, where the
PDZ domain tethers the protein to the cytoskeleton and the LIM
domain or additional internal domains, recruit signaling proteins
to implement corresponding functions.
Assembly of the PDZ- and LIM-domains of the first Enigma isoform identified and characterized (457 amino acids) is as illustrated below [Q9NR12] [2] [11]:
Structure
Shown to the right, the X-ray crystal structure of Enigma PDZ domain has recently been solved to 1.11Å. This revealed a canonical PDZ-domain fold containing six strands and 2 helices.
Function and Interactions
In general, the PDZ domain of Enigma subfamily has been structurally revealed as a classical class I PDZ domain - this is further suggested by its interaction with the C-terminal region of a-actinin-2 [12]. Recently, it has been shown that the PDZ domain of Enigma proteins also interacts with Myotilin and Calsarcin/FATZ C-terminal motifs, which have the characteristics of class III PDZ-binding domain sequences [13]. In addition, experimental evidences indicate Enigma protein may function as a scaffold on which the coordinated assembly of sarcomeric proteins can occur; largely owing to the interactions via its PDZ- and LIM domains with actin-associated proteins of cardiac and skeletal muscle, as well as non-muscle tissues [[[14]]]. It has also been implicated in bone formation and fracture repair[15]. It may also be involved in BMP6 signaling pathway. Generally, it interacts with various PKC isoforms using the LIM domains[16]. The LIM-2 domain has been shown to interact with TBX4, as well as RET in a phosphorylation-independent manner[17].
Pathology
References
- ↑ Faulkner G, Pallavicini A, Formentin E, Comelli A, Ievolella C, Trevisan S, Bortoletto G, Scannapieco P, Salamon M, Mouly V, Valle G, Lanfranchi G. ZASP: a new Z-band alternatively spliced PDZ-motif protein. J Cell Biol. 1999 Jul 26;146(2):465-75. PMID:10427098
- ↑ Zhou Q, Ruiz-Lozano P, Martone ME, Chen J. Cypher, a striated muscle-restricted PDZ and LIM domain-containing protein, binds to alpha-actinin-2 and protein kinase C. J Biol Chem. 1999 Jul 9;274(28):19807-13. PMID:10391924
- ↑ Passier R, Richardson JA, Olson EN. Oracle, a novel PDZ-LIM domain protein expressed in heart and skeletal muscle. Mech Dev. 2000 Apr;92(2):277-84. PMID:10727866
- ↑ Zheng M, Cheng H, Banerjee I, Chen J. ALP/Enigma PDZ-LIM domain proteins in the heart. J Mol Cell Biol. 2010 Apr;2(2):96-102. Epub 2009 Dec 30. PMID:20042479 doi:10.1093/jmcb/mjp038
- ↑ Wang X, Su H. Unraveling enigma in the z-disks. Circ Res. 2010 Aug 6;107(3):321-3. PMID:20689070 doi:10.1161/CIRCRESAHA.110.225615
- ↑ Wu RY, Gill GN. LIM domain recognition of a tyrosine-containing tight turn. J Biol Chem. 1994 Oct 7;269(40):25085-90. PMID:7929196
- ↑ Guy PM, Kenny DA, Gill GN. The PDZ domain of the LIM protein enigma binds to beta-tropomyosin. Mol Biol Cell. 1999 Jun;10(6):1973-84. PMID:10359609
- ↑ Ueki N, Seki N, Yano K, Masuho Y, Saito T, Muramatsu M. Isolation, tissue expression, and chromosomal assignment of a human LIM protein gene, showing homology to rat enigma homologue (ENH). J Hum Genet. 1999;44(4):256-60. PMID:10429367 doi:10.1007/s100380050155
- ↑ Faulkner G, Pallavicini A, Formentin E, Comelli A, Ievolella C, Trevisan S, Bortoletto G, Scannapieco P, Salamon M, Mouly V, Valle G, Lanfranchi G. ZASP: a new Z-band alternatively spliced PDZ-motif protein. J Cell Biol. 1999 Jul 26;146(2):465-75. PMID:10427098
- ↑ Klaavuniemi T, Ylanne J. Zasp/Cypher internal ZM-motif containing fragments are sufficient to co-localize with alpha-actinin--analysis of patient mutations. Exp Cell Res. 2006 May 1;312(8):1299-311. Epub 2006 Feb 14. PMID:16476425 doi:10.1016/j.yexcr.2005.12.036
- ↑ Wu RY, Gill GN. LIM domain recognition of a tyrosine-containing tight turn. J Biol Chem. 1994 Oct 7;269(40):25085-90. PMID:7929196
- ↑ Au Y, Atkinson RA, Guerrini R, Kelly G, Joseph C, Martin SR, Muskett FW, Pallavicini A, Faulkner G, Pastore A. Solution structure of ZASP PDZ domain; implications for sarcomere ultrastructure and enigma family redundancy. Structure. 2004 Apr;12(4):611-22. PMID:15062084 doi:10.1016/j.str.2004.02.019
- ↑ von Nandelstadh P, Ismail M, Gardin C, Suila H, Zara I, Belgrano A, Valle G, Carpen O, Faulkner G. A class III PDZ binding motif in the myotilin and FATZ families binds enigma family proteins: a common link for Z-disc myopathies. Mol Cell Biol. 2009 Feb;29(3):822-34. Epub 2008 Dec 1. PMID:19047374 doi:10.1128/MCB.01454-08
- ↑ Wu RY, Gill GN. LIM domain recognition of a tyrosine-containing tight turn. J Biol Chem. 1994 Oct 7;269(40):25085-90. PMID:7929196
- ↑ Liu Y, Hair GA, Boden SD, Viggeswarapu M, Titus L. Overexpressed LIM mineralization proteins do not require LIM domains to induce bone. J Bone Miner Res. 2002 Mar;17(3):406-14. PMID:11874232
- ↑ Kuroda S, Tokunaga C, Kiyohara Y, Higuchi O, Konishi H, Mizuno K, Gill GN, Kikkawa U. Protein-protein interaction of zinc finger LIM domains with protein kinase C. J Biol Chem. 1996 Dec 6;271(49):31029-32. PMID:8940095
- ↑ Durick K, Gill GN, Taylor SS. Shc and Enigma are both required for mitogenic signaling by Ret/ptc2. Mol Cell Biol. 1998 Apr;18(4):2298-308. PMID:9528800
>