Ferguson ZNF Sandbox
From Proteopedia
| |||||||
| 1znf, 37 NMR models () | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | |||||||
| Non-Standard Residues: | , | ||||||
| |||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
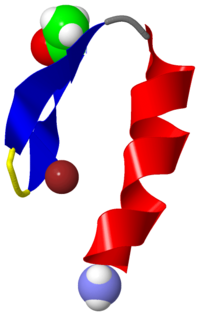
THREE-DIMENSIONAL SOLUTION STRUCTURE OF A SINGLE ZINC FINGER DNA-BINDING DOMAIN
The three-dimensional solution structure of a zinc finger nucleic acid binding motif has been determined by nuclear magnetic resonance (NMR) spectroscopy. Spectra of a synthetic peptide corresponding to a single zinc finger from the Xenopus protein Xfin yielded distance and dihedral angle constraints that were used to generate structures from distance geometry and restrained molecular dynamics calculations. The zinc finger is an independently folded domain with a compact globular structure in which the zinc atom is bound by two cysteine and two histidine ligands. The polypeptide backbone fold consists of a well-defined helix, starting as alpha and ending as 3(10) helix, packed against two beta strands that are arranged in a hairpin structure. A high density of basic and polar amino acid side chains on the exposed face of the helix are probably involved in DNA binding.
About this Structure
1ZNF is a 1 chain structure with sequence from Xenopus laevis. The March 2007 RCSB PDB Molecule of the Month feature on Zinc Fingers by David S. Goodsell is 10.2210/rcsb_pdb/mom_2007_3. Full experimental information is available from OCA.
Reference
- Lee MS, Gippert GP, Soman KV, Case DA, Wright PE. Three-dimensional solution structure of a single zinc finger DNA-binding domain. Science. 1989 Aug 11;245(4918):635-7. PMID:2503871