Introduction
DOPA decarboxylase (Aromatic L-Amino Acid Decarboxylase, tryptophan decarboxylase, 5-hydroxytryptophan decarboxylase, AAAD, or DDC) is an essential lyase enzyme responsible for the conversion (via decarboxylation) of L-3,4-dihydroxyphenylalanine (L-DOPA) and L-5-hydroxytryptophan (5-HTP) to dopamine and serotonin. This 104 kDa protein is a tightly associated α2-dimer that belongs to the aspartate aminotransferase family (fold type 1) of PLP-dependent (vitamin B6-dependent) enzymes, and can be found in abundance in the nervous system, as well as the kidney. Because of its role in the biosynthesis of dopamine, DDC has been utilized in the treatment of Parksinon's disease- a chronic, progressively neurological disorder, thought to be the result of degeneration of dopamine-producing cells in the substantia nigra pars compacta (brain structure in the mesencephalon that plays an important role in reward, addiction, and movement) of the brain.
Structure
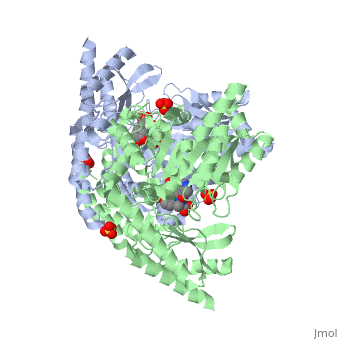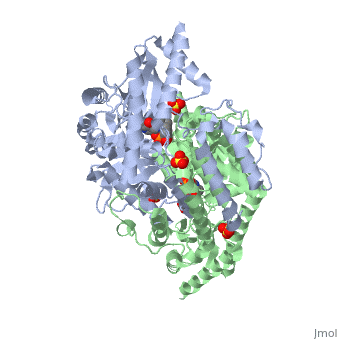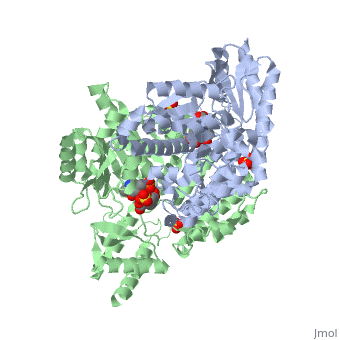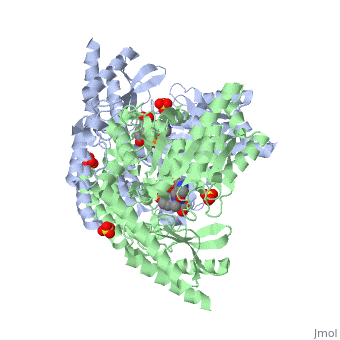
DDC consists of two monomers [SC], each with three distinct domains: the large domain [SC], the C-terminal small domain [SC], and the N-terminal domain [SC]. The consists of the cofactor (PLP- pyridoxal phosphate) binding site and is comprised of a central, seven-stranded mixed β-sheet surrounded by eight α-helices [SC] in a typical α/β fold. The small consists of a four-stranded antiparallel β-sheet with three helices[SC] packed against the face opposite the large domain. The (characteristic of all α-family enzymes) is composed of two parallel helices linked by an extended strand [SC]. This structure flaps over the top of the second subunit and vice versa, with the first helix of one subunit aligning parallel to the equivalent helix of the other subunit, thereby forming the extended dimer interface. Although it is unlikely that this domain represents an autonomous folding unit, it is most likely stable only in the context of the dimer by extending the interface between the two monomers.
Function
The Active Site
DDC's active site is located in a cleft between the two monomer subunits, but is composed mainly of residues from one monomer.The active site is composed of several key residues, including Lys-303, Asp-271, His-192, Thr-82, Ile-101, and Phe-103. In the ligand free form, PLP binds to Lys 303 via a Schiff base linkage. A salt bridge forms between the carboxylate group of Asp 271 and the protonated pyridine nitrogen of PLP yielding a strong electron sink capable of stabilizing the carbanionic intermediates. The only two active site residues from the adjacent monomer, Ile-101 and Phe-103, are part of the substrate binding pocket.
Flexible Loop
Residues 328-338 are a stretch of 11 amino acids that comprise a mobile loop (located near the dimer interface), thought to be significant in the catalytic mechanism. During catalysis, the loop is expected to adhere to a less solvent- and protease-exposed conformation, whereby it loses flexibility and extends toward the active site. Tyr-332 and Lys-334 are highly conserved residues indicative of the flexible loop's possible role in catalysis. The conformational change that occurs is thought to be a result of loop residues directly interacting with the inhibitor. Finally, with such a conformational change, the Tyr-332 residue could then be located closer to the substrate to possibly act as a proton donor for the quinonoid Cα.