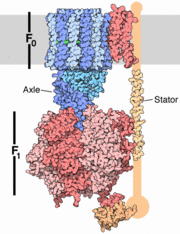
Introduction
The archaeal A1A0 ATP synthase represent a class of chimeric ATPases/synthase , whose function and general structural design share characteristics both with vacuolar V1V0 ATPases and with F1Fo ATP synthases [3]. A1A0 ATP synthase catalyzes the formation of the energy currency ATP by a membrane-embedded electrically-driven motor. The archaeon in this study,Pyrococcushorikoshii OT3 is an anaerobic thermophile residing in oceanic deep sea vents with an optimal growth temperature of 100 degrees C. Anaerobic fermentation is its principle metabolic pathway.
A hyperthermophilic, anaerobic archaeon was isolated from hydrothermal fluid samples obtained at the Okinawa Trough vents in the NE Pacific Ocean, at a depth of 1395m. The strain is obligately heterotrophic, and utilizes complex proteinaceous media (peptone, tryptone, or yeast extract), or a 21-amino-acid mixture supplemented with vitamins, as growth substrates. Sulfur greatly enhances growth. The cells are irregular cocci with a tuft of flagella, growing optimally at 98 degrees C (maximum growth temperature 102 degrees C), but capable of prolonged survival at 105 degrees C.
[4]
The specific enzymatic process in A-ATP synthase reveals novel, exceptional subunit composition and coupling stoichiometries that may reflect the differences in energy-conserving mechanisms as well as adaptation to temperatures at or above 100 degrees C. Because some archaea are rooted close to the origin in the tree of life, these unusual
mechanisms are considered to have developed very early in the history of life and, therefore, may represent the first energy-conserving mechanisms. [1]
Structure of A-ATP synthase catalytic subunit A
Five steps inside the catalytic A-subunit are critical for catalysis. Substrate entrance, phosphate and nucleotide binding, transition-state formation, ATP formation, and product release. The vanadate bound model mimics the transition state. Orthovandate is a useful transition state analog because it can adapt both tetragonal and trigonal bipyramidal coordination geometry. The Avi structure can be compared to the As sulfate bound structure and the Apnp AMP-PNP bound structure. As is analogous to the phosphate binding (substrate) structure and Apnp is analogous to the ATP binding (product) structure[5].
Within the catalytic A subunit there are the N-terminal domain residues 1-79, 110-116, 189-199, the non-homologous region(which region makes this subunit considerably larger than its homologs) residues 117-188, the nucleotide binding alpha-beta domain residues 80-99, 200-437, and C-terminal alpha helical bundle residues 438-588 domains. There are 16 helices and 27 strands.
P-Loop
The is the eight residue consensus sequence of amino acid residues 234-241 GPFGSGKT . The P-loop or phosphate binding loop is conserved only within the A subunits and is a loop preceded by a beta sheet and followed by an alpha helix.
occupies the ADP site. Although not at bonding distances the residues P233 G234 L417 stabilize the first vanadate in the transition state with weak nonpolar interactions. Residues K240 and T241 stabilize with polar interactions.
Residue is a polar serine molecule that interacts with the nucleotides via a hydrogen bond during catalysis. The distance between residue S238 is longest in As, shortest in Avi and intermediate in Apnp . In As a water molecule bridges the gap, which is removed in Avi. Dehydration of the transition state active site is reversed when ATP forms. In Apnp the water molecule interacts with the y-phosphate of ATP.
Comparasons to other known structures
This P-loop has an arched conformation unique to A-ATP Synthase, indicating that the mode of nucleotide binding and the catalytic mechanism is different from that of other syntheses. [6]
The P-loop is unique in archea because it is stable compared to other similar structures. For example, in A-ATP Synthases is involved in P-Loop stabilization.
Another reason why the P-Loop is so stable in A-ATP synthase is because of residue S238. It is significant in the catalytic process in moving towards the y-phosphate of ATP during catalysis, and does this with a hydrogen bond, not a conformational change in the P-loop. In "'F-ATP Synthase"' the homolog to S238 is the non polar A158. Since A158 cannot form hydrogen bonds to interact with the substrate (like s238), the P-loop undergoes a conformational change.[5]