Brittany deRonde/Sandbox 1
From Proteopedia
One of the CBI Molecules being studied in the University of Massachusetts Amherst Chemistry-Biology Interface Program at UMass Amherst and on display at the Molecular Playground.
Contents |
Introduction
HIV Tat, or simply Tat, is a human immunodeficiency virus (HIV) gene that regulates transcription of HIV dsRNA.[1] Tat, which stands for trans-activator of transcription, contains 86 amino acid residues in its sequence.[2][3]
|
Cell Penetrating Peptides
Cell penetrating peptides (CPPs) are proteins with the ability to cross cellular membranes and facilitate the uptake of various cargo, such as small molecules, siRNA, and small DNA fragments.[10] Such cargo can be associated via covalent or non-covalent interactions. Tat is considered a CPP because it contains a protein transduction domain (PTD). PTDs are cation-rich sequences of 10-30 residues, usually containing several Lysine and/or Arginine residues.[11]
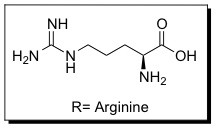
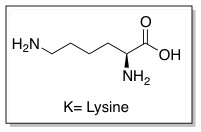
These sequences improve interactions between CPPs and cellular membranes and can help them enter cells. The PTD sequence in Tat is YGRKKRRQRRR (amino acid ), which is Arginine-rich. This 11 amino-acid sequence is now referred to as the TAT peptide, and has be shown to have improved cellular uptake compared to Tat.
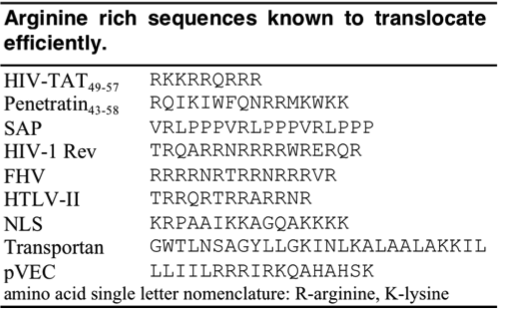
Since the discovery of HIV Tat, other Arginine-rich, natural CPPs have been discovered including, penetratin from Drosophila antennapedia protein, sweet arrow peptide (SAP), HIV-1 Rev, flock house virus (FHV) coat, brome mosaic virus (BMV) Gag, human T-cell lymphotrophic virus (HTLV)-II Rex, and the nuclear localization signal (NLS) from nucleoplasmin. [12][13] The PTD sequences for these proteins can be found in the table below.
Tat-mediated Transduction
The PTD in HIV Tat is highly cationic. When charged residues (Arginine or Lysine) are replaced with neutral residues (Alanine), cellular uptake decreases. It is likely that the cationic charge is necessary for electrostatic interactions with components of the cellular membrane, such as lipid head groups and proteoglycans.[14][15] Other interactions may also play a role in interactions with the cellular membrane, such as hydrophobic and non-covalent interactions.[16] Although cellular uptake is not directly dependent upon architecture (branched vs. linear), it is dependent upon the number of Arginine residues. Polymers with less than five arginine residues are unable to enter cells.[17] As the number of residues increases, cellular uptake improves up until 15 residues where polymers can enter cells but with hindered efficiencies.[18][19]
Transduction
Endocytosis
Drug Delivery Applications
XXXXXX
My Research Interest
As part of the Tew Research Group in the Department of Polymer Science and Engineering at the University of Massachusetts, Amherst, I am interested in the design of Tat-inspired CPPs for drug and gene delivery. Using functionalized oxanorbornene derivatives as monomers, ring opening metathesis polymerization (ROMP) is performed using Grubbs' 3rd generation catalyst to synthesize cationic CPP mimics. By tuning the functionalization of the monomer structures and the resulting CPP molecular weights, cellular uptake and cell viability can be optimized.
References
- ↑ Sodroski et al. Science. 1985. 227, 171-173. [1] http://www.jstor.org/stable/1695050
- ↑ Arya et al. Science 1985. 229, 69-73.
- ↑ Sodroski et al. Science. 1985. 229, 74-77.
- ↑ Dayton et al. Cell. 1986. 44, 941-947.
- ↑ Fisher et al. Nature. 1986. 320, 367-371.
- ↑ Xiao et al. Proc. Natl. Acad. Sci. USA. 2000. 97, 11466-11471.
- ↑ Ensoli et al. Nature. 1990. 345, 84-86.
- ↑ Green et al. Cell. 1988. 55, 1179-1188.
- ↑ Frankel et al. Cell. 1988. 55, 1189-1193.
- ↑ Futaki et al. J. Biol. Chem. 2001. 276, 5836-5840.
- ↑ Wender et al. Adv. Drug Deliv. Rev. 2008. 60, 452-472.
- ↑ Futaki et al. J. Biol. Chem. 2001. 276, 5836-5840.
- ↑ Prochiantz et al. Adv. Drug Deliv. Rev. 2008. 60, 448-451.
- ↑ Ziegler et al. Biochemistry 2003. 42, 9185-9194.
- ↑ Goncalves et al. Biochemistry. 2005. 44, 2692-2702.
- ↑ Ziegler et al. Adv. Drug Deliv. Rev. 2008. 60, 580-597.
- ↑ Wender et al. Proc. Natl. Acad. Sci. 2000. 97, 13003-13008.
- ↑ Mitchell et al. J. Pept. Res. 2000. 56, 318-325.
- ↑ Futaki et al. J. Biol. Chem. 2001. 276, 5836-5840.