We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox207
From Proteopedia
This page is reserved for a work from two students in ESBS (A.Butet and E.Rosati) Thanks.
| |||||||||
| 1gnh, resolution 3.00Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | |||||||||
| |||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
C-reactive protein, CRP
Contents |
CRP, C-reactive Protein
- The C Reactive Protein is a protein of the acute phase, the first described, exclusively synthetized by the liver.
- The CRP was first isolated by Tillett and France in 1930, in patients' serum presenting an acute inflammation. This protein reacted to the polysaccharide C of the pneumocoque, that is where its name comes from. <1>
- The CRP contributes to innate host defense, and plays an important role in inflammatory reactions. It can bind to specific molecules typically exposed during cell death or found on the surfaces of pathogens. It is used as biological marker to reveal an inflammatory reaction and tissue damage. <2>
Structure
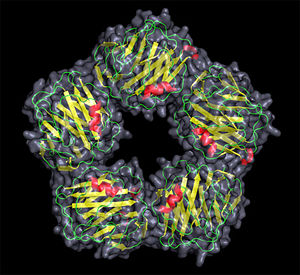
CRP structure <7>
Gene structure, family
- The CRP gene is located on chromosome 1q23. It is composed of two exons and one intron. This gene is regulated by interleukin-6, the principal inducer of the gene during the acute phase. CRP is secreted by hepatocytes.<5>
- The Human CRP belongs to the pentraxin family , proteins having five identical, non-covalently associated subunits that form a symmetrical homopentameric ring. The pentraxin family is highly conserved in evolution.<3>
Size
- Each subunit contains 206 amino acid residues (approximately 23kDa) and is non-glycosylated. The outside diameter of the pentamer is 102 Å, the diameter of the inner core is 30 Å, and the diameter of the protomer is 36 Å. <2>
Detailed strucutre
- Each protomer consists of two anti-parallel β sheets (the lectin fold) with an α helix on the effector face of the protein. The ligand biding site is located on the concave face of the protein, and is composed of loops with 2 calcium ions bound 4 Å apart by protein side-chains.
- The recognition face contains the phosphocholine binding site which consists of two coordinated calcium ions next to a hydrophobic pocket in which the phosphocholine stays.
- There are interprotomer interactions between the subunits: three salt bridges are included and the 115-123 loop of one protomer and the 40-42 and 197-202 regions of adjacent protomers are involved. Moreover, the subunits can rotate by 15-20° around an axis parallel to the central alpha-helix.
Molecular structure and morphology of human CRP. (a) Negatively stained electron micrograph showing the typical pentameric disc-like structure face-on and side-on (arrows). (b) Ribbon diagram of the crystal structure, showing the lectin fold and the two calcium atoms (spheres) in the ligand-binding site of each protomer. (c) Space-filling model of the CRP molecule, showing a single phosphocholine molecule located in the ligand-binding site of each protomer.<3>
- Thanks to this rotation, the alpha-helices can lie closer to the axis of the pentamere, therefore bringing the bound Ca2+ further away from it. On each subunit, we can find phosphocholine bound in a shallow surface pocket. With the help of phosphate groups and Glu81 via the choline moiety (??), the phosphocholine can interact with the two protein-bound ions.
- Moreover, the structure of CRP is different in ill patients. Indeed, in some pathological conditions, the Human CRP is glycosylated. Analysis of the structure showed the systematic absence of two peptide fragments, one at the N-terminus (loop 1-6) in all patients, the other near the C-terminus (loop 189-191) in patients with osteogenic sarcoma and Cushing's syndrome. In a healthy individual, glycosylation sites are inacessible due to the presence of the N-terminal. The loss of these two fragments exposed two potential glycosylation sites on a cleft door. The functional areas of the pentraxin structure remain the same since the Ca2+ and phosphocholine sites are on the opposite site of the pentraxin molecule. <2>
Biomarker
- The CRP concentration is a very useful nonspecific biochemical marker of inflammation and/or tissue damage. It is used since 1977 in the diagnosis and the supervision of the evolution of the infections, because the normalization of its rate is an indication that the infectious phenomenon is mastered. The C Reactive Protein is also a predictive biomarker for cardiovascular disease risk. <4>
- In healthy young adult, the median concentration of CRP is 0.8 mg/l. This value may increase from less than 50 μg/l to more than 500 mg/l (10,000-fold), following an acute-phase stimulus. After a single stimulus, serum concentration of CRP is rising above 5 mg/l in around 6 hours. This increase is proportional to the intensity of the inflammation. The peak is reached during 48 hours. When the stimulus ceases, the circulating CRP concentration falls rapidly. <3>
- There is individual variability in baseline CRP, resulting from non-genetic or genetic factors. Indeed, a polymorphism in the CRP gene intron and promoter has been described, that pertubs expression level.
- The intron of the gene of the CRP is formed of 278 nucleotides, containing a segment of 39 nucleotides rich in GT bases. This segment could be responsible for the formation of the left-handed helix of the Z-form DNA of the CRP. The persons possessing a particular allele combination have a lower concentration of the CRP. It is probably due to structural modifications of the DNA which would affect the transcription of the gene.
- Within the promoter, several polymorphisms were discovered in transcription factor binding E-box sites, what seems to significantly influence the rate of the CRP into the blood.
- This variability should be taken into account when using the CRP as a predictive biomarker. <5>
References
1 <1>
2 <2>
3 <3>
4 <4>
5 <5>
6 <6>
7 <7>

