PPAR-gamma
From Proteopedia
|
Contents |
Introduction
Peroxisome proliferator-activated receptor gamma (γ) is a protein in the nuclear receptors subfamily. It is one of three isotypes (-α, -β/ δ, and -γ) [1] of PPAR receptors and has two protein isoforms governed by splice variations, which result in differences in the length of the amino (N)-terminal region (PPARγ1 and PPARγ2) [2]. PPARγ is involved in transcriptional regulation of glucose and lipid homeostasis [1], and helps regulate adipocyte differentiation [3]. It has a , which allows it to interact with a wide array of ligands. typically triggers a conformational change of PPARγ, notably in the activation function-2 , which aids in the recruitment of co-regulatory factors to regulate gene transcription. PPARγ can form a with retinoic X receptor alpha (RXRα), a process necessary for most PPARγ-DNA interactions [4]. PPARγ is a molecular target for antidiabetic drugs such as thiazolidinediones (TZDs), which makes the protein a target for Type II Diabetes (T2D) drug research. Due to its involvement in metabolic and inflammatory processes, PPARγ also holds potential for treatments of many metabolic and chronic-inflammatory diseases, such as metabolic syndrome and inflammatory bowel disease, respectively. Errors in PPARγ-related regulation have also been implicated in atherosclerosis and various cancers, like colorectal, breast, and prostate cancers.
Overall Structure and Ligand Binding
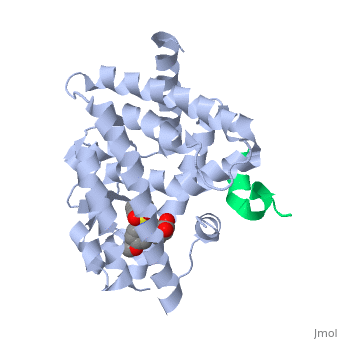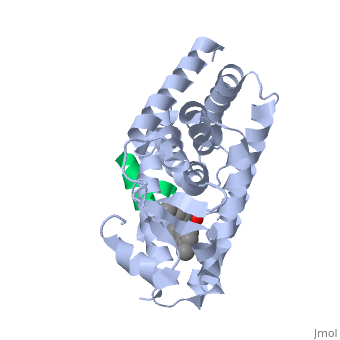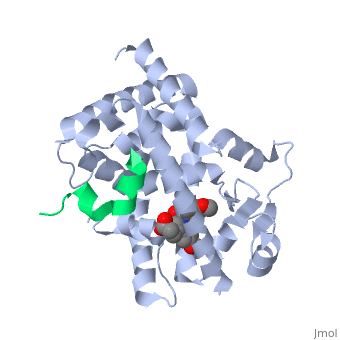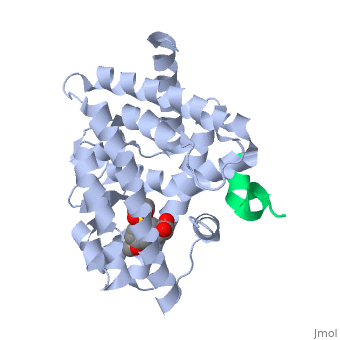
PPARγ is composed of the ligand-independent activation domain (AF-1 region and A/B-domain), a DNA-binding domain (DBD) (C-domain), a hinge region (D-domain), and a ligand-dependent ligand-binding domain (LBD) (E/F-domain and AF-2 region) [5]. The two PPARγ isoforms, PPARγ1 and PPARγ2, differ by only 30 amino acids at the N-terminal end. These added amino acids on PPARγ2 result in increased potency and adipose-selectivity, which makes this protein a key player of adipocyte differentiation [3]. The is composed of 13 α helices and 4 short β strands [1]. It has a T-shaped binding pocket with a volume of ~1440 Å3 [1, 6], which is larger than that of most nuclear receptors [7], allowing for interactions with a variety of ligands [8]. The PPARγ LBD is folded into a helical sandwich to provide a binding site for ligands. It is located at the C-terminal end of PPARγ and is composed of about 250 amino acids [5]. Activation by full agonists occurs through hydrogen bond interactions between the S289, H323, Y473, and H449 residues of the PPARγ-LBD [7] and polar functional groups on the ligand which are typically carbonyl or carboxyl oxygen atoms. Agonist binding results in a conformational change of the LBD AF-2 region, which is necessary for coactivator recruitment. This change can either be dramatic or subtle [1], which leads to stabilization of a charge clamp between helices H3 and H12 [9] to aid in associations with the LXXLL (L, leucine; X, any amino acid) motif of the coactivator [1, 10]. Ligand binding of PPARγ is regulated by communication between the N-terminal A/B domain, which is adjacent to the DBD, and the carboxyl-terminal LBD [11].
|
Ligand Activity
PPARγ ligands, fall into one of three categories: full agonist, partial agonist, or antagonist [5]. Full agonists have higher efficacy for activating PPARγ and higher potency [7], and their binding leads to the more dramatic conformational change [1]. Binding of partial agonists leads to the more subtle change [1] and results in lower efficacy and potency [7]. Antagonists do not activate PPARγ, so there is either no conformational change to exclude coactivators or a minor conformational change to accommodate corepressors [9,5]. Natural ligands of PPARγ include fatty acids, eicosanoids, and prostaglandins [4-8,11-13].
Coactivators/Corepressors
The of PPARγ is a groove created by hydrophobic residues of the H3, H3’, H4, and H12 helices [1]. Stabilization of the AF-2 domain is important for coactivator interactions, and is achieved through ligand binding [1]. Upon agonist binding, coactivators and other chromatin-remodeling cofactors, like histone deacetylases, are recruited and transcription is activated [14]. Coactivators can be regulated at the transcriptional and post-transcriptional levels, as well as by protein-kinase cascades [3]. PPARγ can actively silence genes it is bound to by recruiting a corepressor in the absence of a ligand. Once this occurs, an antagonist binds to stabilize the AF-2 region, preventing interactions with coactivators and activation of transcription [9]. Corepressor binding creates a three-turn α-helix corepressor motif important for preventing the AF-2 domain from assuming an active conformation [9]. Common coactivators of PPARγ include CBP/p300, the SRC family, and TRAP220 [3]. Common corepressors include SMART, NCoR, and RIP140 [3].
PPARγ/RXRα
PPARγ shows preferential heterodimerization with RXRα [3]. The asymmetry of PPARγ/RXRα packs positively and negatively charged regions together, and is needed for PPARγ binding to DNA [1]. The LBD and DBD of PPARγ are located close together, whereas the RXRα LBD and DBD are positioned farther apart. This difference in region proximity plays a role in heterodimerization [5]. The PPARγ/RXRα complex associates with PPAR response elements (PPREs) in promoter regions of targeted genes [8]. Each ligand-bound PPAR/RXRα complex will bind to a specific PPRE based on the recruited cofactor [8].
Functions
PPARγ is a key regulator of glucose and lipid homeostasis [1]. PPARγ mediates adipocyte differentiation and alters insulin sensitivity, inflammatory processes, and cell proliferation [11]. Ligand-dependent mechanisms include inhibition of inflammatory cytokine production and macrophage activation [8]. In addition to adipocytes, PPARγ is also found in the retina, cells of the immune system, and colonic epithelial cells [5]. PPARγ controls the expression of various genes, particularly those involved in fatty acid metabolism [5]. Regulated genes include adipocyte fatty acid binding protein (aP2), lipoprotein lipase (LPL), and acyl-CoA oxidase [4]. Tissue-specific deletions of PPARγ lead to insulin resistance, low viability of mature adipocytes, and lipodystrophy [3].
Potential of PPARγ in Disease Treatment
Synthetic TZDs were the first class of PPARγ ligands identified [8]. The most potent and selective agonist of PPARγ from this class is the insulin sensitizer rosiglitazone [8]. T2D is linked to higher levels of free fatty acids (FFAs) and triglycerides in the blood [8], which is a contributing factor for insulin resistance. Treatment with rosiglitazone and other TZDs reduces levels of FFAs and triglycerides, helping to restore insulin sensitivity [8]. TZDs also work by increasing levels of GLUT-4 and decreasing levels of pro-inflammatory cytokines [5]. PPARγ is found in high levels in colonic epithelial cells. The role of PPARγ in these cells may be related to regulation of immune response and colon inflammation [12]. The onset of Inflammatory Bowel Disease is thought to be caused by inflammatory cytokines present in the colon [12]. In patients with ulcerative colitis, colonic epithelial cells displayed impaired expression of PPARγ, an important mediator of aminosalicylate activities in Inflammatory Bowel Diseases [13]. TZD ligands could be implemented to reduce colonic inflammation [12]. Agonists have also been used in the treatment of colitis and psoriasis by inhibiting the inflammatory response of the epithelium and reducing cytokine production [8]. PPARγ inhibits activity of nuclear factor NFκB, which is higher in active ulcerative colitis patients [15]. PPARγ could also be implemented in the treatment of other chronic inflammation-related diseases. Immunomodulatory effects have been found with PPARγ agonists [16]. Rosiglitazone alongside adiponectin reduces renal disease, atherosclerosis, and production of autoantibodies, all of which are characteristic of the inflammatory autoimmune disease Systemic Lupus Erythematosus (SLE) [16]. PPARγ ligands hold potential as cancer treatments [11] due to their ability to inhibit angiogenesis, the process required for the growth and metastasis of solid tumors [8]. PPARγ activators have pro-differentiation and anti-proliferation effects [3]. TZDs have also been shown to inhibit proliferation of human breast, prostate, and colon cancer cells [8].
3D structures of PPAR
Peroxisome Proliferator-Activated Receptors
Additional Resources
For additional information See: Diabetes
For additional information See: Regulation of Gene Expression
For additional information See: Peroxisome Proliferator-Activated Receptors
References
[1] Gampe Jr RT, Montana VG, Lambert MH, et al. Asymmetry in the PPARγ/RXRα crystal structure reveals the molecular basis of heterodimerization among nuclear receptors (2000) Molecular Cell, 15(9), pp.545-555.
[2] Zieleniak A, Wójcik M, Woźniak LA. Structure and physiology functions of the human peroxisome proliferator-activated receptor γ (2008) Arch. Immunol. Ther. Exp., 56 (5), pp. 331-345.
[3] Tontonoz P, Spiegelman BM. Fat and Beyond: The Diverse Biology of PPARγ (2008) Annu. Rev. Biochem., 77, pp. 289-312.
[4] Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: Nuclear control of metabolism (1999) Endocrine Reviews, 20 (5), pp. 649-688.
[5] Lewis SN, Bassaganya-Riera J, Bevan DR. Virtual Screening as a Technique for PPAR Modulatory Discovery (2010) PPAR Research, 2010, pp. 861238.
[6] Itoh T, Fairall L, Amin K, et al. Structural basis for the activation of PPARγ by oxidized fatty acids (2008) Nature Structural and Molecular Biology, 15 (9), pp.924-931.
[7] Pochetti G, Godio C, Mitro N, et al. Insights into the mechanism of partial agonism: crystal structures of the peroxisome proliferator-activated receptor γ ligand-binding domain in the complex with two enantiomeric ligands (2007) Journal of Biological Chemistry, 282 (23), pp.17314-17324.
[8] Murphy GJ, Holder JC. PPAR-γ agonists: Therapeutic role in diabetes, inflammation and cancer (2000) Trends in Pharmacological Sciences, 21 (12), pp. 469-474.
[9] Xu HE, Stanley TB, Montana VG, et al. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARα (2002) Nature, 415 (6873), pp.813-817.
[10] Kallenberger BC, Love, JD, Chatterjee VKK, Schwabe JWR. A dynamic mechanism of nuclear receptor activation and its perturbation in a human disease (2003) Nature, 10 (2), pp.136-140.
[11] Shao D, Rangwala SM, Bailey ST, Krakow SA, Reginato MJ, Lazar MA. Interdomain communication regulating ligand binding by PPARγ (1998) Nature, 396, pp. 377-380.
[12] Su CG, Wen X, Bailey ST, Jiang W, Rangwala SM, Keilbaugh SA, Flanigan A, Murthy S, Lazar MA, Wu GD. A Novel therapy for colitis utilizing PPAR-γ ligands to inhibit the epithelial inflammatory response (1999) J Clin Invest., 104(4), pp. 383-389.
[13] Dubuquoy L, Rousseaux C, Thuru X, Peyrin-Biroulet L, Romano O, Chavatte P, Chamaillard M, Desreumaux P. PPARγ as a new therapeutic target in inflammatory bowel disease (2006) International Journal of Gastroenterology and Hepatology, 55 (9), pp.1341-1349.
[14] McKenna NJ, O'Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators (2002) Cell, 108 (4), pp. 465-474.
[15] Sartor, RB. Mechanisms of disease: pathogenesis of Crohn’s disease and ulcerative colitis (2006) Nature, 3(7), pp. 390-407.
[16] Aprahamian T, Bonegio RG, Richez C, Yasuda K, Chiang L, Sato K, Walsh K, Rifkin IR. The Peroxisome Proliferator-Activated Receptor γ Agonist Rosiglitazone Ameliorates Murine Lupus by Induction of Adiponectin (2009) the Journal of Immunology, 182, pp. 340 -346.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Lera Brannan, David Canner, Alexander Berchansky