Sandbox Reserved 433
From Proteopedia
| This Sandbox is Reserved from January 19, 2016, through August 31, 2016 for use for Proteopedia Team Projects by the class Chemistry 423 Biochemistry for Chemists taught by Lynmarie K Thompson at University of Massachusetts Amherst, USA. This reservation includes Sandbox Reserved 425 through Sandbox Reserved 439. |
Contents |
Caspase 3- 1rhk
Introduction
Caspases are a group of dimeric cysteine proteases that play important roles to control the ultimate steps of apoptosis and innate inflammation; they are also very important in cellular development, homeostasis and in a wide range of diseases such as neurodegeneration, ischemia and cancers. During apoptosis, the initiator caspases (caspases 8 and 9) as upstream regulators cleave and activate with the downstream executioner caspases (caspases 3, 6 and 7). Then the activated executioner caspases will cleave upwards of 500 key proteins and DNA, which finally cause the death of cells.[1]
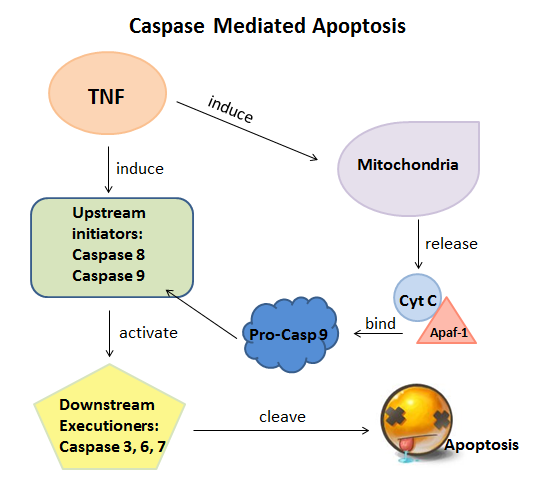
|
There are many caspase structures are researched, and most of them involve either peptide, protein inhibitors or unattractive candidates for drug development.[2] is one of the downstream executioner caspases which interacts with caspase 8 and 9. It is formed from a 32 kDa zymogen that is cleaved into 17 kDa and 12 kDa subunits. It contains four anti-parallel beta sheets from p17 and two from p12, which will come together to make a heterodimer that will interact with another heterodimer to form a total 12-stranded structure surrounded by that is owned by caspases only.[3][4] As an executioner caspase, the caspase-3 zymogen does not have activity until it is cleaved by an initiator caspase after apoptotic signaling events have occurred.[5] Caspase 3 has many of the typical characteristics which all currently-known caspases also own. For instance, its contains acysteine residue, and histidine residue, . Also, Caspase 3 is able to be activated by diverse death-inducing signals, which includes the chemotherapeutic agents. Pathways of the activation of Caspase 3 was already identified that were either dependent on or independent of mitochondrial cytochrome c release and Caspase 9 function. It also plays an significant role many other apoptotic scenarios in tissue-, cell type-, or death stimulus-specific manner.[6]
Since Caspase 3 cleaves a wide range of cellular substrates including structural proteins and DNA repair enzymes and also activates an endonuclease caspase-activated DNAse, which causes the DNA fragmentation that is characteristic of apoptosis, it has many biological functions, such as normal brain development and several significant diseases including Alzheimer’s disease, Polycystic Kidney Disease and Cancers. In a very frequently-occurring cancer, breast cancer, Caspase 3 acts as a key mediator whose mRNA and protein expression was examined, which leads that the rates of apoptosis as measured by both caspase 3 activation and nucleosome release are higher in breast cancer than in nonmalignant breast tissue.[7] In another cancer, lung cancer, researches shown that polymorphisms in the caspase 3 gene may influence caspse 3 production and activity, thereby modulating the susceptibility to lung cancer.[8] Thus, to cure cancers, studying on Caspase 3 becomes a significant entrance.
Overall Structure
|
The structure shown here is
While the heterodimer shown above and below is the active complex; the original of casepase-3 will form a heterotetramer through interaction with another caspase-3 at the final beta-strand in chain B. This forms a containing a non-polar twelve stranded beta sheet surrounded by polar non-polar alpha-helixes which is a structural characteristic of caspases particularly caspases-1, 3, 7, and 8.[9]
Caspase 3, as shown above and below, is composed of three chains (referred to as A (p17), B (p12), and C) that are 147, 102, and 5 amino acids long (respectively). The three chains are primarily held together through a parallel non-polar beta-strand connection between chains A and B and an anti-parallel polar beta-strand connection between chains B and C. Chain A consists of 7 beta-strands and 4 alpha helixes. Chain B has 4 beta-strands and 3 alpha helixes. Only chains A and B are conserved. Chain C consists of a single beta-sheet which can be of variable length and sequence which, though close to the active site on chain A does not seem to be part of it. This is why, though there are three chains shown here, Caspase-3 is considered a heterodimer, and not a heterotrimer.
Binding Interactions
|
The of the enzyme has been determined to be Cys-285 and His-237. His-237 stablizes the peptide while Cys-285 attacks cleaving the peptide bond. Cys-285 and Gly 238 stabilize the transition state through hydrogen bond and therefore also very important. In addition there is a to stabilize the Asp residue of the peptide undergoing cleavage. The pocket consists of polar amino acids such as Arg, Ser, and Gln which allows polar Asp to be held in the correct orientation for the reaction to occur [4].
Since the catalytic site of the molecule has been determined, inhibitors have been developed which have the ability to bind to these essential amino acids to prevent the enzyme from working. Caspase 3 is regulated with small peptides and other proteins in vivo but since proteins are often large and need specific conditions to work, they are often not ideal to use for medical treatments.
There are numerous inhibitors available commericially as well as research for more specific and easily controlled inhibitors. Ac-DEVD-aldehyde is a commercially available inhibitor of Caspase 3. The binds directly with the Cys 285 of the active site leading to the formation of a hemithioacetal and stablization by hydrogen bonding with Cys 285 and Gly 238 [5] This inhibits Caspase 3 through competative inhibition.
Other molecules such as nicotinic acid aldehydes have also been seen to inhibit Caspase 3 and have shown great promise due to peptide-less characteristics. In experimental trials nicotinic acid aldhyde A had a similar as Ac-DEVD-aldehyde binding directly to the catalytic diad and preventing substrate binding. However there are other nicotinic acid aldhydes that have and have shown to have greater inhibition. For example it has strong interactions with Tyr 388 and Phe381 which lead researchers to believe that these amino acids also have an important role in the catalytic mechanism of the enzyme. This reinforces the idea that there are many parts of the protein that are vital for correct function and shape, not exclusively the active sites [5].
After creating a list of for multiple inhibitors, it can been seen that the most important region of binding is around the catalytic diyad.
Additional Features
|
Role in apoptosis
1. How it is activated (green scene of activation ligand?)
2. catalytic site (if binding interactions doesn't cover this)
Potential role in stem-cell differentiation:
In addition to its role in the caspase cascade of apoptosis, caspase-3 also plays a role in stem cell differentiation, specifically with neural cells. This seems rather counter-intuitive, as stem cell differentiation leads to cell proliferation and apoptosis is cell death. However, as it turns out, cells undergoing differentiation have similar cytoskeleton changes, as well as membrane fusion and fission events as cells undergoing apoptosis. [2]
"rather than by simply limiting self-renewal. In this model, caspase-3 may simultaneously engage factors to promote the gene expression profile and resulting phenotypic changes that result in a specific differentiated cell type" (Abdul-Ghani and Megeney 515)
"Cells undergoing either apoptosis or differentiation display comparable cytoskeletal rearrangements, membrane fusion, and fission events as well as similar alterations in chromatin and nuclear architecture" (Abdul-Ghani and Megeney 515)
"Taken together, our data implicate signaling proteins that mediate cytoskeletal remodeling in the response to nonapoptotic caspase-3 activity" (Fernando, Brunette, and Megeney 7)
"The persistent levels of nestin in caspase-3-inhibited cells suggest that the initiation of neurogenesis is delayed. Although nestin levels do not increase within the caspase-3-inhibited groups, the amount of nestin relative to the control group is greater. This may suggest that neural precursor cells that do not undergo differentiation-induced apoptosis remain alive." (Fernando, Brunette, and Megeney 7)
"increased caspase-3 activity leads to the activation of cellular remodeling kinases such as PAK1 and ASK1. In turn, these kinases also activate downstream protein kinases such as p38 and are known to target the nucleus and subsequently phosphorylate differentiation-specific transcription factors." (Fernando, Brunette, and Megeney 7)
Another article: [1]
Not sure what to do for green scenes here though...
Credits
Introduction - Di Lin
Overall Structure - Austin Virtue
Drug Binding Site - Jill Moore
Additional Features - Alex Way
References
- ↑ Jeanne A. Hardy and James A. Wells, Dissecting an Allosteric Switch in Caspase-7 Using Chemical and Mutational Probes, THE JOURNAL OF BIOLOGICAL CHEMISTRY,VOL. 284, NO. 38, pp. 26063–26069, September 18, 2009.http://www.jbc.org/content/284/38/26063.short
- ↑ Crystal structure of the complex of caspase-3 with a phenyl-propyl-ketone inhibitorReducing the peptidyl features of caspase-3 inhibitors: a structural analysis.Becker, J.W., Rotonda, J., Soisson, S.M., Aspiotis, R., Bayly, C., Francoeur, S., Gallant, M., Garcia-Calvo, M., Giroux, A., Grimm, E., Han, Y., McKay, D., Nicholson, D.W., Peterson, E., Renaud, J., Roy, S., Thornberry, N., Zamboni, R.,Journal: (2004) J.Med.Chem. 47: 2466-2474
- ↑ Salvesen GS (January 2002). "Caspases: opening the boxes and interpreting the arrows". Cell Death Differ. 9 (1): 3–5. doi:10.1038/sj.cdd.4400963. PMID 11803369.
- ↑ Lavrik IN, Golks A, Krammer PH (October 2005). "Caspases: pharmacological manipulation of cell death". J. Clin. Invest. 115 (10): 2665–72. doi:10.1172/JCI26252. PMC 1236692. PMID 16200200
- ↑ Walters J, Pop C, Scott FL, et al. (December 2009). "A constitutively active and uninhibitable caspase-3 zymogen efficiently induces apoptosis". Biochem. J. 424 (3): 335–45. doi:10.1042/BJ20090825. PMC 2805924. PMID 19788411
- ↑ Emerging roles of caspase-3 in apoptosis,Alan G Portera and Reiner U Jänicke,Institute of Molecular and Cell Biology, The National University of Singapore, 30 Medical Drive, Singapore 117609, Republic of Singapore: http://www.nature.com/cdd/journal/v6/n2/abs/4400476a.html
- ↑ Caspase 3 in breast cancer.,O'Donovan N, Crown J, Stunell H, Hill AD, McDermott E, O'Higgins N, Duffy MJ. Source, Department of Medical Oncology, St. Vincent's University Hospital, University College Dublin, Dublin 4, Ireland. http://www.ncbi.nlm.nih.gov/pubmed/12576443
- ↑ Identification of Polymorphisms in the Caspase-3 Gene and Their Association With Lung Cancer Risk Jin Sung Jang,Kyung Mee Kim,Jin Eun Choi,Sung Ick Cha,Chang Ho Kim,Won Kee Lee,Sin Kam,Tae Hoon Jung and Jae Yong Park, Department of Biochemistry, School of Medicine, Kyungpook National University, Daegu, Korea. http://onlinelibrary.wiley.com/doi/10.1002/mc.20397/pdf
- ↑ Salvesen GS "Caspases: opening the boxes and interpreting the arrows" Nature Publishing Group 2002 vol:9 iss:1 pg:3 -5
3. Abdul-Ghani, Mohammad, and Megeney, Lynn A. "Rehabilitation of a Contract Killer: Caspase-3 Directs Stem Cell Differentiation". Cell Stem Cell, Vol. 2, No. 6 (2008): 515-16. Web. 04 Apr 2012. [2]
4. Fernando, Pasan, Brunette, Steve, and Megeney, Lynn A. "Neural Stem Cell Differentiation is Dependent Upon Endogenous Caspase-3 Activity". The Journal of the Federation of American Societies For Experimental Biology, Vol. 19 (2005): 1671-3. Web. 04 Apr 2012.[3]
5. Cohen G. M. (1997). Caspases: the executioners of apoptosis. Biochem. J. 326(Pt 1), 1–16.
6. Becker JW, Rotonda J, Soisson SM, Aspiotis R, Bayly C, Francoeur S, Gallant M, Garcia-Calvo M, Giroux A, Grimm E, Han Y, McKay D, Nicholson DW, Peterson E, Renaud J, Roy S, Thornberry N, Zamboni R. Reducing the peptidyl features of caspase-3 inhibitors: a structural analysis. J Med Chem. 2004;47:2466–2474.
7. Salvesen GS "Caspases: opening the boxes and interpreting the arrows" Nature Publishing Group 2002 vol:9 iss:1 pg:3 -5
