User:Marvin O'Neal/Antibody OspA and OspB
From Proteopedia
Contents |
Introduction
Borrelia burgdoferi
The causative agent of Lyme disease is Borellia burgdorferi, a spirochaete found in the gut of hard bodied ticks of genus Ixodes. A factor contributing to the severity of Lyme disease is its resistance to certain forms of complement-dependent immune response by the evasion of the alternative complement pathway and the blocking of complement C3. [1] This resistance increases the importance of the complement independent immune response when combating B. burgdorferi. Certain fragment antigen binding regions (fab) of IgG and IgM monoclonal antibodies (mAbs)are bactericidal even in the absence of complement. Binding of these fabs to their corresponding outer surface protein (OspA and OspB) of B. burgdoferi leads to the lysis of the bacteria.[2]
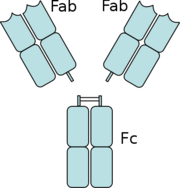
Fragment Antigen Binding (fab)
Fab consists of a heavy chain and light chain and each chain is composed of a variable and a constant region. The paratope is located in the N terminal of the variable region of the heavy and light chains of the fab. H6831 and CB2 are IgG mAbs that targets the C-terminal of OspB and LA-2 is an IgM mAb that targets the C-terminal of OspA. [3]
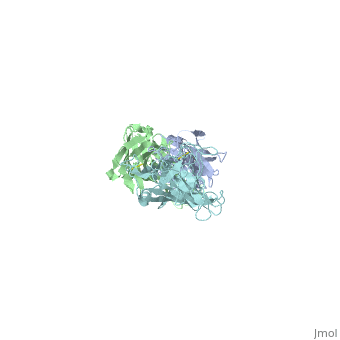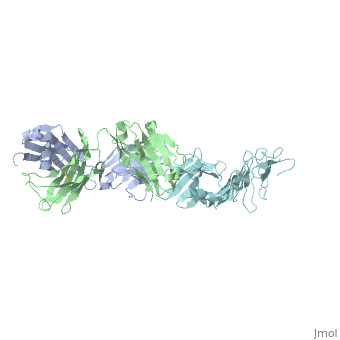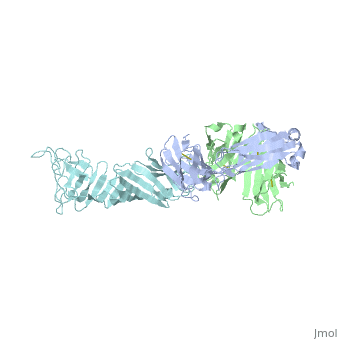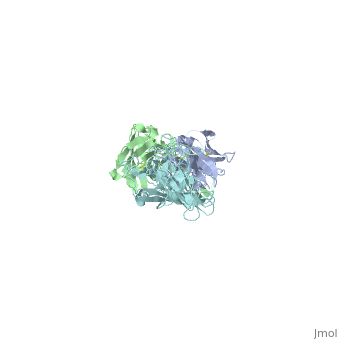
OspB and H6831
| |||||||
: ·· ·· |
Interaction between OspB and H6831
The consist of two components, the outer surface protein , and the , subdivided into the and the . Most hydrogen bonds and electrostatic interactions that are responsible for the binding of H6831 to OspB are between the at the C-terminal of OspB and some that include tyrosine, tryptophan, glutamate, and histidine.
The majority of hydrogen bonds and electrostatic interactions are between loop 2 (residues 250-254) and the fab heavy chain. in loop 2 of OspB has a necessary and major role due to its central position in the exposed loops. A mutation at its position abrogates the binding interaction and causes the resistance of the bacteria to the bactericidal effect of the fab. Lys 253 binds to the two aromatic residues on the fab heavy chain, tyrosine and tryptophan. It also makes hydrogen bonds with the glutamate in the heavy chain of the fab and forms an ionic bond. Carbonyl in loop 1 (green link) of the OspB interacts with histidine in the fab heavy chain. of OspB interacts with fab light chain. [4].
Structural changes to OspB in the complexed form
The binding of H6831 to OspB leads to some conformational changes in OspB compared to its free form. Study, done by becker using crystallography, has showed that the most significant difference is the loss of the central , strands 1-4. Both small positional shifts near the Fab binding site and a few larger structural changes away from the binding site were observed. The largest shifts (7– 8 Å) correspond to the repositioning of a loop opposite the Fab-binding site (residues 218 –220). In the free OspB structure, all regions that exhibit shifts are adjacent to the central sheet; in the OspB-H6831 complex they all shift toward, and slightly overlap, the position of the missing sheet. These observations suggest that the larger conformational changes are related to the loss of the central sheet, which could have happened through proteolytic cleavage or fortuitous crystal contacts [4].
Bactericidal action
The fab binding destabilizes the outer membrane (OM) of B. burdorferi, with subsequent formation of spheroplasts. It has been observed that the bactericidal action, but not the binding, requires the presence of divalent cations (Mg2+ and Ca2+). Escudero et al. study demonstrated the inability of fab to kill bacteria in the absence of the divalent cations. It was speculated that OspB-Cb2 (a fab similar to H6831) complexes could create physical openings in the OM allowing for rapid infusion of electrolytes, increasing the osmolarity of the periplasm, and leading to the lysis of the cell [5].
OspA and LA-2
|
OspA
OspA is 53% similar to OspB. OspA is usually undetectable in the early stages of lyme disease, because it is down regulated when OspC is expressed (provide citation). Despite the similarity between OspA and OspB, both in vivo and vitro OspB is susceptible to cleavage by exogenous proteases, whereas OspA is relatively resistant (Becker, structural). This characteristic might have contributed to OspA being a proper vaccine to lyme disease that is used to prevent transmission of B.burgodorferi from tick vector to mammalian host by complement-independent killing.[6]
Interaction between OspA and LA-2
LA-2 is an IgM murine monoclonal antibody that interacts with three exposed loop on the C-terminal of OspA. These interactions include eight direct hydrogen bonds, four solvent-bridged hydrogen bonds, three ion pairs, and numerous van der Waals interactions [6].
Structural changes to OspA in the complexed form
Conformational changes upon the binding of OspA and LA-2 show that LA-2 recognition of OspA involves an induced fit mechanism where loops 1-3 conformations shift to optimize complementarity to the antigen-combining site [6]. The overall structure of the C-terminal of OspA is unchanged upon the binding of LA-2 with comparison to the free OspA. The maximum atomic shift is 4.7Å at the site of Ser206.[7]
- ↑ LaRocca TJ, Benach JL. The important and diverse roles of antibodies in the host response to Borrelia infections. Curr Top Microbiol Immunol. 2008;319:63-103. PMID:18080415
- ↑ Connolly SE, Benach JL. The versatile roles of antibodies in Borrelia infections. Nat Rev Microbiol. 2005 May;3(5):411-20. PMID:15864264 doi:10.1038/nrmicro1149
- ↑ Putnam FW, Liu YS, Low TL. Primary structure of a human IgA1 immunoglobulin. IV. Streptococcal IgA1 protease, digestion, Fab and Fc fragments, and the complete amino acid sequence of the alpha 1 heavy chain. J Biol Chem. 1979 Apr 25;254(8):2865-74. PMID:107164
- ↑ 4.0 4.1 Becker M, Bunikis J, Lade BD, Dunn JJ, Barbour AG, Lawson CL. Structural investigation of Borrelia burgdorferi OspB, a bactericidal Fab target. J Biol Chem. 2005 Apr 29;280(17):17363-70. Epub 2005 Feb 15. PMID:15713683 doi:10.1074/jbc.M412842200
- ↑ Escudero; Halluska et al. 1997
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedding - ↑ Ding W, Huang X, Yang X, Dunn JJ, Luft BJ, Koide S, Lawson CL. Structural identification of a key protective B-cell epitope in Lyme disease antigen OspA. J Mol Biol. 2000 Oct 6;302(5):1153-64. PMID:11183781 doi:10.1006/jmbi.2000.4119
Proteopedia Page Contributors and Editors (what is this?)
Christopher Smilios, Philip J. Pipitone, Safa Abdelhakim, Alexandros Konstantinidis, Jaime Prilusky