Sandbox Reserved 472
From Proteopedia
| This Sandbox is Reserved from 13/03/2012, through 01/06/2012 for use in the course "Proteins and Molecular Mechanisms" taught by Robert B. Rose at the North Carolina State University, Raleigh, NC USA. This reservation includes Sandbox Reserved 451 through Sandbox Reserved 500. | ||||||
To get started:
More help: Help:Editing For more help, look at this link: http://www.proteopedia.org/wiki/index.php/Help:Getting_Started_in_Proteopedia Renin
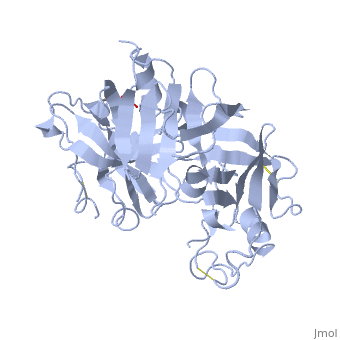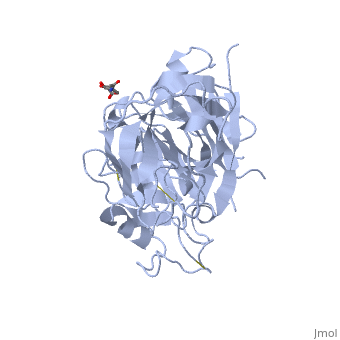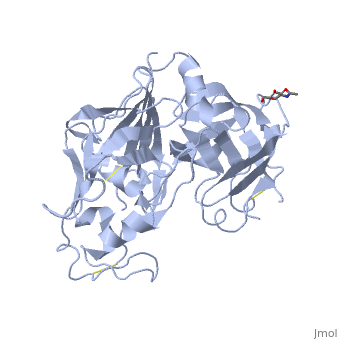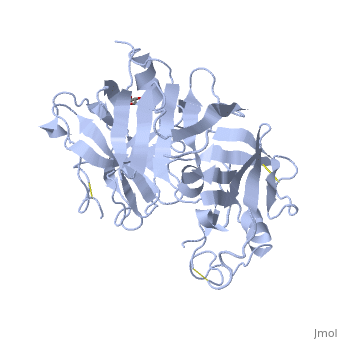
IntroductionRenin was first discovered in 1898 by physiologist Robert Tigerstedt and his medical student Bergman. It is an aspartyl-protease that plays a major role in the physiological rennin-angiotensin-aldosterone system; therefore indirectly involved in homeostatic control of salt, volume and blood pressure. The enzymatic reaction of renin is the cleavage of Lue bond in angiotensinogen to make angiotensin. In the human body, renin is synthesized by juxtaglomerular epitheloid cells proceeding stimulation by the prostaglandins and sympathetic nerves. Secretion of renin is controlled by three secondary messengers: cAMP, cGMP, and cytosolic Calcium. Studies have shown that the protease is inhibited by angiotensin II, high blood pressure, salt and volume overload; all through negative feedback loops. Once produced, renin enters the body’s circulation in renal system through afferent arterioles. Expression of the renin gene in these cells have been heavily studied in humans and mice and mapped to chromosome 1. StructureAs displayed to the right of the page, a folded recombinant human renin molecule consists of two lobes. Located between these lobes is the at the bottom of the cleft consisting of two catalytic aspartic residues. At this site, the pro-segment of the inactive form of renin forms a plug thereby hindering substrate binding. The of the protease include seven helices containing 35 residues and 28 strands containing the remaining 136 residues. X-ray diffraction was the method of choice used to solve the structure. |