P53-DNA Recognition
From Proteopedia
| This Sandbox is Reserved from 22 April 2012, through 31 August 2012 for use in the course "Protein DNA" taught by Remo_Rohs at the La Canada High School, USA. This reservation includes Sandbox Reserved 169 through Sandbox Reserved 170. |
To get started:
More help: Help:Editing |
This is a joint project of La Canada High School and University of Southern California students, mentored by Professor Remo Rohs.
Contents |
A Base Pairing Variant Enhances p53 Binding to a Response Element
Introduction and Biological Role of the Tumor Suppressor p53
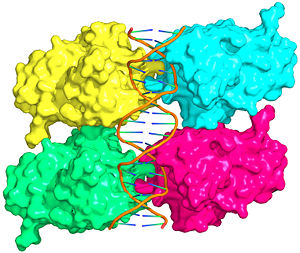
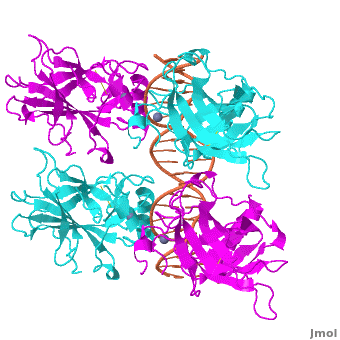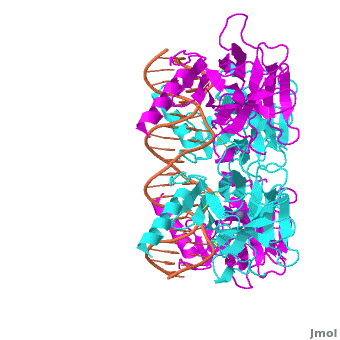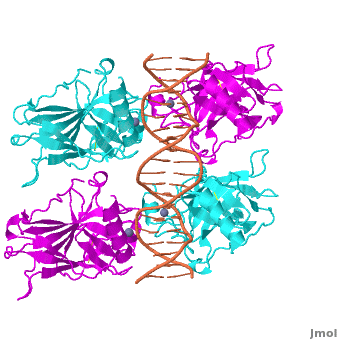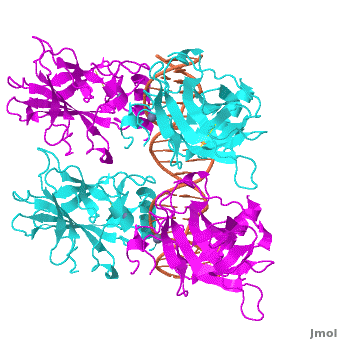
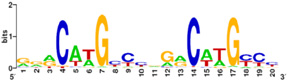
Also known as the Guardian of the Genome, the tumor suppressor p53 is central in the natural defense against human cancer. The protein is activated by stress factors that can compromise the genomic integrity of the cell, and this activation unleashes the function of p53 as transcription factor. It binds as a tetramer (Figure 1) to a large range of DNA response elements. The p53 consensus site (Figure 2) is formed by two decameric half-sites, each containing a core element (red), that are separated by a variable number of base pairs (blue).
Binding of p53 to different response elements leads to distinct biological responses, such as cell-cycle arrest, senescence, or apoptosis. These different pathways correspond, at least in part, to differences in p53-DNA binding affinity and stability, which are determined by specific protein-DNA interactions.
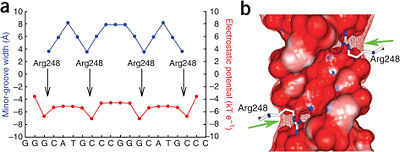
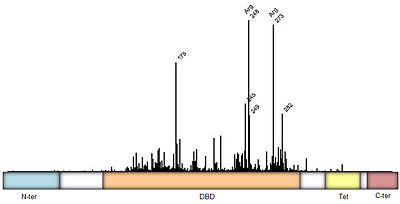
Mutations of p53 residues are associated with 50% of human cancers. Such mutations are predominantly located in the p53-DNA binding domain (DBD) based on an analysis of human tumors (Figure 3). Particularly, arginine residues in the p53-DNA interface were found in tumors with high frequencies.
Structural Description of p53-DNA Complex
Domain Architecture and Tetramerization
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
Remo Rohs, Eric Martz, Alexander Berchansky, Julia Tam, Sharon Kim, Bailey Holmes, Angel Herraez, Joseph M. Steinberger, Eran Hodis, Masha Karelina, Michal Harel, Ana Carolina Dantas Machado, Jaime Prilusky, Skyler Saleebyan, Joel L. Sussman, Keziah Kim